Back to Journals » OncoTargets and Therapy » Volume 11
miRNA-30c can be used as a target in the diagnosis and treatment of osteosarcoma
Authors Sun R, Muheremu A , Hu Y
Received 23 July 2018
Accepted for publication 25 November 2018
Published 14 December 2018 Volume 2018:11 Pages 9091—9099
DOI https://doi.org/10.2147/OTT.S181177
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Rongxin Sun,1,2 Aikeremujiang Muheremu,3 Yihe Hu1
1Department of Orthopedics, Xiangya Hospital, CentralSouth University, Kaifu District, Changsha, Hunan 86410008, China; 2Department of Orthopedics, Sixth Affiliated Hospital of Xinjiang Medical University, Tianshan District, Urumqi, Xinjiang 86830001, China; 3Department of Spine Surgery, Sixth Affiliated Hospital of Xinjiang Medical University, Tianshan District, Urumqi, Xinjiang 86830001, China
Objective: Osteosarcoma is a highly malignant osseous sarcoma with poor prognosis. Previous studies indicated that miRNA-30c may play an important role in the development of osteosarcoma, but its mechanism is not yet clear. The current research was carried out to explore the potential applications of miR-30c in the diagnosis and treatment of osteosarcoma.
Materials and methods: Real-time PCR and in situ hybridization were used to test the correlation between miR-30c and the onset of osteosarcoma. In vitro transfection of miR-30 mimic was used to test the effect of miR-30c on the development of osteosarcoma. Cell Counting Kit-8, formation of Petri dish clones, in vivo formation of tumor, flow cytometry tests and Transwell analysis were used to assess the effect of miR-330c on the metastatic potential and invasiveness of osteosarcoma.
Results: Reverse transcriptase-PCR analysis and in situ hybridization tests revealed that the expression of miR-30c was lower in the osteosarcoma tissue than in normal bone tissue (P<0.05). Low expression of miR-30c was associated with advanced osteosarcoma staging and low cellular differentiation. Multivariate analysis revealed that lower expression of miR-30c was associated with shorter survival of patients (P<0.01). U2OS cell growth was significantly inhibited when transfected with miR-30c mimic. Flow cytometry analysis revealed that overexpression of miR-30c could induce apoptosis of osteosarcoma cells. In vitro Petri dish cloning experiment showed that overexpression of miR-30c reduced the cloning ratio of U2OS cells from 21% to 7%. At the same time, overexpression of miR-30c inhibited the formation of sarcoma in nude mice. Transwell experiments indicated that overexpression of miR-30c could reduce the invasiveness of U2OS cells.
Conclusion: Low expression of miR-30c was associated with high probability of onset and aggressiveness of osteosarcoma and shorter patient survival. Upregulation of miR-30c could downregulate the invasiveness of osteosarcoma. Therefore, miR-30 can be used in the development of future diagnostic and therapeutic techniques.
Keywords: miRNA-30c, osteosarcoma, diagnosis, invasiveness, treatment
Introduction
Osteosarcoma is one of the most common malignant lesions of the bone. It constitutes about 20% of all primary malignant osseous sarcomas. Osteosarcoma is one of the most fatal malignancies due to its characteristics such as rapid growth and early migration to vital organs.1 Despite the application of in-block tumor resection and new adjuvant chemotherapy, the 5-year survival of osteosarcoma patients without lung metastasis is limited to 60%–70%.2 Patients with lung metastasis, however, have a 5-year survival of <30%.3
Early diagnosis and treatment is the key for achieving satisfactory prognosis. Early diagnosis of osteosarcoma can be facilitated by noninvasive tests of specific tumor markers. Advancements in the research of miRs provided clinicians with novel targets for the diagnosis and treatment of osteosarcoma. miR is a type of noncoding RNA with 19–25 nucleotides. It may participate in a series of biochemical processes such as transcription regulation, DNA methylation and histone acetylation;4,5 among these, DNA methylation and histone acetylation are closely related to the development and formation of cancer in mammals.
Normal physiological functions of miRs include regulating the signal pathways of cell differentiation, proliferation, metabolism and apoptosis. In malignant lesions, expression of certain miRNAs is up- or downregulated.6 Previous research has proven that miR-30c plays an important role in the proliferation and aggressiveness of tumor cells.7 Xia et al reported that the expression of miR-30c was significantly decreased in lung cancer cells, and that knocking out miR-30c can significantly increase the aggressive behavior of lung cancer A549 cells.8 Huang et al found that anoxia can decrease the expression of miR-30c and increase patient susceptibility for renal cancer.9 Sanchez-Diaz et al19 used high-flux miRNA chips and found that miR-30c is downregulated in patients with osteosarcoma.10
According to these findings, we have hypothesized that miR-30c plays an important role in the growth and metastasis of osteosarcoma. This study was designed and carried out to find the possible mechanism on how miR-30c affects the biological behavior of osteosarcoma, and provide a novel target for the diagnosis and treatment of osteosarcoma.
Materials and methods
Patients and specimens
All the human and animal studies in the current research were approved by the ethics committee of the Sixth Affiliated Hospital of Xinjiang Medical University. All patients gave oral or signed consent for application of their clinical materials for research purposes. This study was conducted in accordance with the Declaration of Helsinki. All the animal studies were performed following the guidelines and regulations of Xinjiang Medical University.
Specimens of osteosarcoma patients who were treated in the Sixth Affiliated Hospital of Xinjiang Medical University between January 2007 and December 2016 were consecutively included in the primary analysis. The patients received no chemotherapy or radiotherapy at the time of specimen extraction. After the specimen was extracted, it was immediately stored at −80°C in a freezer and moved to liquid nitrogen. All the patients received outpatient follow-up visits. Physical checkups and computed tomography scans were carried out at each follow-up.
Quantitative reverse transcriptase-PCR (qRT-PCR) assay of the specimens
According to the user manual of Thermo Fisher Scientific for Trizol, RNA was extracted from the specimens. U6 was used as the internal control. Primers of U6 and miR-30c were designed and chemosynthesized by Shanghai Jima Biotech Ltd (Shanghai, China). Quantitative PCR template was obtained by reverse transcribing with an miR reverse transcription kit. Primers used for qRT-PCR are: forward: 5′-GTcGTATccAGTGcAGGGTccGAGGTATTcGcAcTGGATAcGAc-3′, reverse: 5′-GcAAcccGTAGATccGAAcTT-3′. RT-PCR was applied to test the expression of miR-30c in osseous tumors and normal bone tissue.
Osteosarcoma in situ hybridization technique
The tumor tissue was embedded in paraffin; 4 μm slices of the tumor tissue were prepared and mounted on immunohistochemical specific glasses loaded with electric charge, and in situ hybridization was performed according to the manual of Exiqon (Vedbæk, Denmark). Spectrum analysis by Nuance™ multi-spectrum microscopic imaging system was used to test the expression of miR-30c in the test subjects. Two senior pathologists independently analyzed the slides and picked one or two typical fields under 200 times enlarged view. Positive staining of the cells was studied using the inForm analytical software. Using the criteria given in Tables 1 and 2, visual fields in each tissue sample were analyzed and the amount of positive staining was calculated.
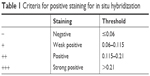  | Table 1 Criteria for positive staining for in situ hybridization |
  | Table 2 Number of colonies formed by different cell typesa |
Cell culturing and transfection
Normal hFOB1.19 cells and U-2OS osteosarcoma cells were obtained from the cell bank of Chinese Academy of Sciences. All the cells were cultured using DMEM high-glucose culture medium containing 10% BSA, 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C in a 5% CO2 incubator. Cells in a good condition and in the exponential growth state were extracted and used for further analysis.
Transfection and grouping of osteosarcoma cells
U2OS cells in the exponential growth state were extracted, mixed with miR-30c mimics or scramble mimics and liposome 2000 into a volume of 600 μL, mixed evenly by blowing with Transferpettor and kept at 25°C for 1 hour. Three hundred microliters of the mixture were added to a six-pore plate, horizontally rocked by the oscillator and cultivated for 4 hours. Then, the culture medium was changed with the transfection medium. After being cultivated for 48 hours under 25°C and trypsinization, the cells were recollected for further analysis.
miR-30c mimics were obtained from Ambion Corporation (Austin, TX, USA). Cells were divided into three groups: 1) control group: cultured under normal circumstances; 2) negative control group: scramble mimics + liposome 2000 were added in the transfection medium and 3) experimental group: miR-30c mimics + liposome 2000 were added in the transfection medium.
Cell cloning
Cells were extracted when they were in the exponential growth sate, washed in PBS twice, digested in 0.25% trypsin for 2 minutes and lightly mixed with Transferpettor to form single cell suspension. Cell concentration of the suspension was adjusted to 1×106/mL.
Five milliliters of 0.7% agarose were mixed evenly with 5 mL of 2× DMEM culture medium in an aseptic test tube. Two hundred microliters of the cell suspension with 1×106/mL cell concentration were added to the test tube, mixed evenly, injected on a Petri dish coated with 1.2% agarose, and thus, two agarose layers were formed. After the top layer was solidified, the cells were cultured for 2 weeks under normal circumstances. Cell clones were counted under an inverted microscope. The clone forming ratio was calculated as (number of clones/number of inoculated cells) ×100%.
Cell vitality test (Cell Counting Kit [CCK]-8 method)
Twenty-four hours after culturing, cells were inoculated in 96-well cell culture plates at a concentration of 1,000 cells/well. Cells from one well were extracted on days 1, 2, 3 and 4 after the inoculation and added into the reaction solution made by mixing culture medium and CCK-9 at a ratio of 9:1. Several wells with only the reaction solution were used as control. The 96-well plates were extracted after being incubated for 1–2 hours. ELISA was used to test the OD value of each well under 450 nm. Cell proliferation ratio was calculated by the following formula: relative proliferation ratio (%) = (average OD value of experimental wells − average OD value of zero control wells)/(average OD value of control wells − average OD value of zero control wells) ×100%. The experiment was carried out three times and the average value was considered the final value.
Testing the effect of different treatment methods on U2OS cell apoptosis
Single-cell suspensions of U2OS cells in control, negative control and experimental groups at 48 hours after transfection were centrifuged under 4°C, 1,000 rpm for 5 minutes. The cell concentration was adjusted to 1×106/mL. Cells were stained following the manual of Annexin V/propidium iodide kit. Flow cytometry tests were used to assess the apoptosis of cells. Each experiment was repeated three times.
Assessment of cell migration and intrusion abilities (Transwell method)
Twenty microliters of Matrigel substrate primer (diluted to 1:8) were laid on Transwell top chamber and incubated at 37°C for 60 minutes until it solidified and formed propria layer. DMEM 20% serum was added in 24-well plate at a concentration of 600 μL in each plate. Cells were collected after being trypsinized and were suspended in high-glucose DMEM with no serum. The average density of the cells was 10,000/mL. Cell suspension was added to the Transwell cell culture chamber at a concentration of 200 μL in each chamber. The chamber was immersed in 24-well cell culture chamber and incubated at 37°C in a 5% CO2 incubator for 24 hours. Transwell chambers were extracted, washed in PBS and the filter was fixed in methanol for 10 minutes. The fixation fluid was discarded and an appropriate volume of crystal violet solution was added for 5 minutes. Q-tips were used to scratch the cells on the filter membrane on the Transwell top chamber. The filter membranes were encapsulated and were observed under a microscope. The invasion ability of the cells was quantitated by counting the cells in five different areas under a microscope.
Subcutaneous tumor formation experiment in nude mice
Nude mice used in the current research were healthy male BALB/c type mice. Thirty mice were randomly assigned to three groups, with ten mice in each group. Three groups of cells (vacant control, control and experimental) were cultured until they reached exponential growth state. A 1 mL syringe was used to extract the cells and inject them percutaneously in the mice (0.2 mL [1×106 cells] in each mouse). The mice were fed in conventional circumstances; their growth, diet, weight and tumor formation were observed daily. The volume and weight of tumor were measured once each week. After the length (L) and width (W) of the tumor were measured, the volume of tumors was calculated by the following formula: V = L × W × W/2. Animals were sacrificed by dislocation of cervical spine in deep anesthesia 5 weeks after the injection. The complete tumor was extracted; fat tissues were cleaned, weighed and prepared for further analysis.
Statistical analysis
All the data were analyzed using IBM SPSS 22.0 software (IBM Corporation, Armonk, NY, USA). The pathological parameters were compared by chi-squared analysis. Kaplan–Meier analysis was used for survival analysis. Cox regression analysis was used for single and multiple variant analyses. One-way ANOVA was used to compare the parameters among the groups. The difference was considered significant when P-value was <0.05.
Results
A total of 37 patients (20 male, 17 female; average age: 19.7±2.4 years, range 11–64 years) were included in the initial specimen analysis. Diameter of the tumor was <10 cm in 15 patients and >10 cm in 22 patients. Histological staging of osteosarcoma tissues was made according to the 2002 WHO osseous sarcoma tissue classification (second edition). Clinical tumor staging was made according to the classification of Enneking et al,20 there were 2 patients with stage I, 21 patients with stage II and 14 patients with stage III lesion. Lung metastasis was found in 25 of these patients. The average follow-up time was 38.6 months (range 6–48 months). Normal bone specimens of 29 patients (19 male, 10 female; average age: 18.3±1.6 years, range 13–52) who were treated for bone fracture at the same period were included as controls. There were no significant differences between the two groups concerning the gender and age of the patients (P>0.05).
Expression of miR-30c in osteosarcoma lesions
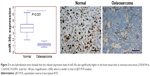
qRT-PCR analysis revealed that the expression of miR-30c in the osteosarcoma lesions was significantly lesser than that in normal bone tissue (Figure 1). The expression of miR-30c was significantly different between the normal and osseous sarcoma cells (one-way ANOVA, P<0.01). In situ hybridization tests showed that miR-30c was mainly located in the nucleus of the osteosarcoma cells. In the meanwhile, it was strongly positive in both nucleus and cytoplasm in the normal bone cells (Figure 2).
Relationship between the levels of miR-30c and patient outcome
Assigning the median value (3.72) of miR-30c expression in in situ hybridization test as a cutoff, 37 patients were divided into two groups. Twenty-six patients were included in the low-expression group and eleven patients in the high-expression group. Table 3 describes the relationship between the levels of miR-30c and the clinical characteristics of patients. Cox regression analysis revealed that there was no significant relationship between the expression of miR-30c and clinical characteristics of osteosarcoma patients such as patient age, type of tissue, location and size of the tumor and tumor metastasis.
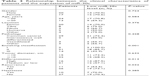  | Table 3 Relationship between the clinical characteristics of patients and the expression of miR-30c |
Relationship between the expression of miR-30c and patient prognosis
Single and multiple variant Kaplan–Meier survival analyses were used to determine the relationship between the expression of miR-30c and patient prognosis (Table 4). Kaplan–Meier survival curve showed that low expression of miR-30c was positively related to shorter time of survival (Figure 3). Cox model analysis showed that low expression levels of miR-30c (HR 23.74, CI 1.58–52.66, P<0.001), late Enneking staging (HR 19.94, CI 1.27–42.85, P<0.001) and low differentiation of tumor tissues (HR 16.23, CI 1.05–39.28, P=0.008) were positively related to shorter survival. After excluding other factors by multiple variant Cox regression analysis, the level of miR-30c was found to be a prognostic factor of osteosarcoma (Table 5).
  | Table 4 Low expression level of miR-30c and the prognosis of osteosarcoma patients |
  | Figure 3 Kaplan–Meier analysis of osteosarcoma patients: low expression level of miR-30c is closely related to the short survival of patients (P<0.01). |
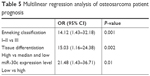  | Table 5 Multilinear regression analysis of osteosarcoma patient prognosis |
Effect of miR-30c on U2OS cell clone formation
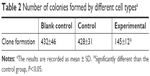
Clone formation experiment was carried out to assess the proliferation potential of single cells. In the current research, compared to the scramble mimics control group and the blank control group, cells in the miR-30c mimics experimental group formed significantly lesser colonies (P<0.05; Figure 4). There were no significant differences between the scramble mimics control group and the blank control group (Figure 4; Table 2).
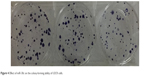  | Figure 4 Effect of miR-30c on the colony-forming ability of U2OS cells. |
Effect of miR-30c on the proliferation of U2OS cells
In the current research, CCK-8 curve was used to show the proliferation of U2OS cells. After the transfection of miR-30c mimics, the proliferation ratio of cells significantly decreased within 72 hours. The proliferation capacity of cells was significantly lower in the experimental group than in the control group at 72, 96 and 120 hours after transfection (P<0.05; Figure 5).
  | Figure 5 The proliferation capacity of cells was significantly lower in the experimental group than in the control group at 72, 96 and 120 hours after transfection (*P<0.05). |
Effect of miR-30c on the apoptosis of U2OS cells
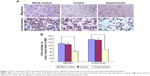
Flow cytometry analysis showed that the ratio of apoptosis was 2.94%±0.78%, 3.02%±1.03% and 15.04%±2.37% in blank control group, control group and experimental group. One-way ANOVA indicated that there were no significant differences between the blank control group and the control group (P>0.05), but there was a significant difference between the experimental group and other groups (P<0.01; Figure 6). This indicates that transfection of miR-30c can promote apoptosis of U2OS cells.
Effect of miR-30c on the migration and invasiveness of U2OS cells
After transfecting U2OS cells with miR-30c mimics, their migration and invasiveness were found to be significantly inhibited (Figure 7A and B).
Effect of miR-30c on the volume of tumors formed after percutaneously injecting U2OS cells
The difference was significant among the groups on comparing the weight and volume of percutaneously formed tumors (Figure 8; Table 6).
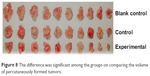  | Figure 8 The difference was significant among the groups on comparing the volume of percutaneously formed tumors. |
  | Table 6 Weight and volume of the tumors in the three groups of nude micea |
Discussion
In the early stages of osteosarcoma, the clinical symptoms are always concealed; therefore, most patients are already at the late stages of osteosarcoma when diagnosed. Recent investigations on miRs have shown that miR-30c participates in the proliferation, differentiation and apoptosis of cells.11 Its expression is found to be downregulated in malignancies such as bladder cancer, invasive micropapillary carcinoma, as well as neurosarcoma.12,13 miR-30c was found to inhibit miRNA and reduce the proliferation and migration of cancer cells, making them more sensitive to chemotherapy. miR-30c was also reported to be a predictive factor in the prognosis of estrogen receptor–positive breast cancer patients undergoing endocrinology treatment.14
However, there is no sufficient data on the current literature on the effect of miR-30c on osteosarcoma and its potential applications. In the current research, in order to avoid error that could be caused by the method of measurement, we used both qRT-PCR and in situ hybridization methods. Both methods consistently showed that the level of miR-30c was significantly high in the normal bone tissue than in osteosarcoma tissue. Moreover, the low expression of miR-30c was associated with more invasive behavior of osteosarcoma cells. This is in accordance with the findings with other types of cancers such as breast cancer, endometrial carcinoma, prostate cancer and ovarian cancer. The current research has also shown that low level of miR-30c is positively related to lower differentiation of tumor cells and higher clinical grading, which implies that patients with low miR-30c levels tend to have unfavorable prognosis.
The working mechanism of miR-30c on osteoporosis could include several pathways. Recent studies have found miR-30c to be a target for repressing tumor cell vitality; for example, miR-30c downregulates MTA1 gene in endometrial carcinoma cells and BCL9 gene in ovarian cancer cells.15,16 Cytoskeleton controlling protein Twinfilin-1 (Twf1) and Vimentin are related to neoplasm invasiveness, and miR-30c can downregulate the expression of Twf1 and Vimentin. Immunohistochemical studies showed that miR-30c can directly combine with MTA1 cancer gene, indicating that miR-30c can affect the avidity of cancer cells by inhibiting MTA1 gene.8 miR-30c was also reported to inhibit the growth of breast cancer cells by inhibiting KRAS signal pathway. In the study of Dobson et al, transfection of miR-30c in breast cancer MDA-MB-231 cells was found to downregulate the nephroblastoma overexpressed gene (NOV) and reduce its invasiveness.17 These mechanisms may also play a role in the miR-30c-based treatment of osseous sarcoma. Further studies are needed to confirm these hypotheses.
Recurrence and metastasis are the main prognostic factors for patients with osteosarcoma. Fang et al18 reported that miR-30c could significantly inhibit the proliferation, differentiation and metastasis of breast cancer cells. They reported that miR-30 could significantly inhibit tumor formation of DU145 prostate cancer cells in nude mice. In the current research, when U2OS osteosarcoma cells were transferred with miR-30c mimics, proliferation, in vitro clone formation, antiapoptosis and migration abilities of the cells were significantly inhibited. In the in vivo experiment, tumor formation capabilities of U2OS osteosarcoma cells transferred with miR-30c mimics were significantly inhibited. All these results indicate that miR-30c can be used as a target in future clinical research.
Disclosure
The authors report no conflicts of interest in this work.
References
Portugal Gomes AC, Marques Ribeiro Naccarato C, Marchiori E. Osteosarcoma: lymphatic spread in the thorax. Arch Bronconeumol. 2016;52(9):486. | ||
Pileczki V, Cojocneanu-Petric R, Maralani M, Neagoe IB, Sandulescu R. MicroRNAs as regulators of apoptosis mechanisms in cancer. Clujul Med. 2016;89(1):50–55. | ||
Wu XQ, Dai Y, Yang Y, Huang C, Meng XM, Wu BM, Li J. Emerging role of microRNAs in regulating macrophage activation and polarization in immune response and inflammation. Immunology. 2016;148(3):237–248. | ||
Liu K, Sun X, Zhang Y, Liu L, Yuan Q. miR-598: a tumor suppressor with biomarker significance in osteosarcoma. Life Sci. 2017;188:141–148. | ||
Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122–1143. | ||
Heinzelmann J, Unrein A, Wickmann U, et al. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: a comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21(3):1046–1054. | ||
Chen P, Wang H, Yang F, Chen H, He W, Wang J. Curcumin promotes osteosarcoma cell death by activating miR-125a/ERRα signal pathway. J Cell Biochem. 2017;118(1):74–81. | ||
Xia Y, Chen Q, Zhong Z, Xu C, Wu C, Liu B, Chen Y. Down-regulation of miR-30c promotes the invasion of non-small cell lung cancer by targeting MTA1. Cell Physiol Biochem. 2013;32(2):476–485. | ||
Huang J, Yao X, Zhang J, et al. Hypoxia-induced downregulation of miR-30c promotes epithelial–mesenchymal transition in human renal cell carcinoma. Cancer Sci. 2013;104(12):1609–1617. | ||
Li X, Jiang H, Xiao L, Wang S, Zheng J. miR-200bc/429 inhibits osteosarcoma cell proliferation and invasion by targeting PMP22. Med Sci Monit. 2017;23:1001–1008. | ||
Widodo, Djati MS, Rifa’i M. Role of microRNAs in carcinogenesis that potential for biomarker of endometrial cancer. Ann Med Surg (Lond). 2016;7:9–13. | ||
Ma Y, Gong J, Liu Y, Guo W, Jin B, Wang X, Chen L. MicroRNA-30c promotes natural killer cell cytotoxicity via up-regulating the expression level of NKG2D. Life Sci. 2016;151:174–181. | ||
Li S, Yang C, Zhai L, et al. Deep sequencing reveals small RNA characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res Treat. 2012;136(1):77–87. | ||
Presneau N, Eskandarpour M, Shemais T, Henderson S, Halai D, Tirabosco R, Flanagan AM. MicroRNA profiling of peripheral nerve sheath tumours identifies miR-29c as a tumour suppressor gene involved in tumour progression. Br J Cancer. 2013;108(4):964–972. | ||
Rodríguez-González FG, Sieuwerts AM, Smid M, et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;127(1):43–51. | ||
Zhou H, Xu X, Xun Q, et al. MicroRNA-30c negatively regulates endometrial cancer cells by targeting metastasis-associated gene-1. Oncol Rep. 2012;27(3):807–812. | ||
Dobson JR, Taipaleenmäki H, Hu YJ, et al. hsa-mir-30c promotes the invasive phenotype of metastatic breast cancer cells by targeting NOV/CCN3. Cancer Cell Int. 2014;14(1):73. | ||
Fang Y, Shen H, Cao Y, et al. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res. 2014;47(1):60–69. | ||
Sanchez-Diaz PC, Hsiao TH, Zou Y, et al. In silico functional analyses and discovery of survival-associated microRNA signatures in pediatric osteosarcoma. Oncoscience. 2014;1(9):599–608. | ||
Enneking WF, Spanier SS, Goodman MA. A system for the surgical staging of musculoskeletal sarcoma. Clin Orthop Relat Res. 1980;(153):106–120. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.