Back to Journals » OncoTargets and Therapy » Volume 11
miRNA-19 promotes non-small-cell lung cancer cell proliferation via inhibiting CBX7 expression
Received 25 July 2018
Accepted for publication 12 September 2018
Published 7 December 2018 Volume 2018:11 Pages 8865—8874
DOI https://doi.org/10.2147/OTT.S181433
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjeev K. Srivastava
Xiaogang Peng, Li Guan, Baoan Gao
Department of Respiratory, China Three Gorges University, Yichang Central People’s Hospital, Yichang City, Hubei Province, China
Background: miR-19 is a critical carcinogenic miRNA that participates in important biological processes of human malignancies. CBX7 plays a key role in lung cancer development and progression. In the present study, for the first time, we investigated the correlation between miR-19 and CBX7 in non-small-cell lung cancer (NSCLC).
Methods: miR-19 expression in NSCLC tissues and lung cancer cell lines was detected using quantitative reverse transcriptase PCR (qRT-PCR). Luciferase reporter assay, qRT-PCR, Western blot, and immunohistochemical assay were conducted to identify the target reaction of miR-19 and CBX7. Moreover, the influence of miR-19 on lung cancer cell proliferation, migration, and invasion was studied including cell counting kit-8 assay, scratch assay, transwell assay, flow cytometry assay, and staining assays.
Results: miR-19 was overexpressed in NSCLC tissues and lung cancer cell lines. Luciferase reporter assay demonstrated that miR-19 could inhibit CBX7 expression via binding to the 3'-UTR of CBX7. Furthermore, miR-19 remarkably decreased CBX7 protein and mRNA expression. Additionally, overexpression of miR-19 could significantly enhance lung cancer cell proliferation and migration.
Conclusion: miR-19 functions as a tumor accelerator promoting lung cancer cell proliferation through targeting CBX7 and inhibiting its expression.
Keywords: miR-19, NSCLC, CBX7, proliferation
Introduction
Lung cancer is identified as one of the most common and insidious malignancies with high morbidity and mortality rates worldwide, accounting for approximately 13% of total cancer deaths.1–3 The lack of systematic diagnosis and effective treatment results in a higher annual incidence rate and a lower 5-year survival rate of patients.4,5 Furthermore, a considerable number of patients are diagnosed at an advanced stage of lung cancer and thus could not receive timely treatment.6 Therefore, lung cancer has attracted increasing global concerns in recent years. The molecular mechanisms of tumor genesis of lung cancer urgently need to be elucidated in order to develop more effective treatment strategies. miRNAs are short noncoding RNA molecules containing 20–22 nucleotides, and induce degradation or inhibit translation of target genes through binding to the 3′-UTR of mRNAs.7–9 miRNAs functionally participate in a series of biological processes including cell proliferation, differentiation, apoptosis, and metabolism under normal physiological conditions, whereas the aberrant expression of miRNAs may cause adverse events including induction of oncogenic pathways.10,11 Extensive investigations have demonstrated that miRNAs play crucial roles in cancer formation, progression, and metastasis including lung cancer.12–14 Yang et al reported that miR-564 functions as a tumor suppressor in human lung cancer by targeting ZIC3.15 Tian et al stated that miR-21 overexpression predicts advanced clinicopathological features and poor prognosis in patients with non-small-cell lung cancer (NSCLC).16 Hence, the correlation between miRNAs and lung cancer should be studied in order to investigate the tumor pathogenesis.
miR-19, a member of miR-17–92 cluster, has been identified as a critical carcinogenic miRNA associated with biological processes of human malignancies including lymphomas, breast cancer, gastric cancer, and glioma by regulating target genes and related signaling pathways.17,18 Olive et al confirmed that miR-19 promotes B-cell lymphoma proliferation by inhibiting PTEN directly and activating AKT/mTOR pathway.19 Liu et al reported that miR-19 inhibits the growth of glioma cells by negative regulation of PTEN.20 Recently, several literature have reported that CBX7 plays a critical role in the initiation and progression of human tumors, including gastric cancer, lymphoma, and prostate cancer, by acting as an oncogene or a tumor suppressor.21–23 Pallante et al reported that the loss of CBX7 expression predicts a more advanced stage of neoplastic disease and a poor survival rate.24 However, the correlation between miR-19 and CBX7 has not been studied so far. In the present research, for the first time, we confirm that miR-19 promotes lung cancer cell proliferation by targeting CBX7.
Materials and methods
Subjects and NSCLC tissues
For this study, 15 NSCLC tissue samples and 15 non-tumor samples were harvested from patients at China Three Gorges University, Yichang Central People’s Hospital. The non-tumor samples were obtained by surgically resecting tissues at a distance of more than 5 cm from the tumors. All the samples were properly stored prior to further assay. Besides, according to tumor diameters and presence or absence of metastasis, the 15 NSCLC tissues were divided into two groups for immunohistochemical assay: tumors less than 5 cm in diameter without metastasis and tumors more than 5 cm in diameter with metastasis. The patients received no treatment before surgery. For all the samples, clinicopathological information (smoking, age, pathological subtype, TNM classification, tumor stage, lymph node stage, and differentiation status) was available. The research was approved by the Ethics Committee of China Three Gorges University, Yichang Central People’s Hospital, and all the subjects voluntarily provided signed informed consent.
Cell lines
Human lung cancer cell lines including NCL-H460, NCL-H1975, 95D, HCL-H358, A549, and NCL-H1299 and control cell line BEAS-2B were purchased from Institute of Biochemistry and Cell Biology, CAS (Shanghai, China). All cells were cultured in RPMI-1640 medium containing with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C with 5% CO2 in a humidified condition.
Transfection
A549 cells were divided into four groups: untransfected cells, negative control miRNA mimic (miR-NC)-transfected cells, miR-19-transfected cells, and miR-19+ CBX7-transfected cells. The cells were transfected using Lipofectamine™ 2000 (Thermo Fisher Scientific). miR-19 expression level was detected using quantitative reverse transcriptase PCR (qRT-PCR) after 48 hours of transfection.
RNA extraction and qRT-PCR
For RNA detection, total RNA was isolated from tissues and cells using Trizol (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. All the tissues were properly homogenized to increase extraction efficiency. Furthermore, RNA was reverse-transcribed using One Step PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Kusatsu, Japan) and quantitatively analyzed using ABI 7500 QPCR (Thermo Fisher Scientific) based on manufacturer’s recommendations. Relative gene expression was detected using 2−ΔΔCt method with β-actin and U6 SnRNA as internal references for mRNA and miRNA, respectively.
Luciferase reporter assay
CBX7 3′-UTR luciferase plasmid, named as pMIR-CBX7-wt, and the mutated CBX7 3′-UTR luciferase vector, named as pMIR-CBX7-mut, were separately constructed and co-transfected with miR-19 mimic or miR-NC into A549 cells, which were divided into four groups (pMIR-CBX7-wt+ miR-19 mimic, pMIR-CBX7-wt+ miR-NC, pMIR-CBX7-mut+ miR-19 mimic, and pMIR-CBX7-mut+ miR-NC), according to the instructions mentioned in Lipofectamine™ 2000 kit (Thermo Fisher Scientific). After incubation for 36 hours in a cell incubator, the cells were lysed with passive lysis buffer and the relative luciferase activity of different groups was tested with Renilla luciferase as internal reference.
Protein extraction and western blot
A549 cells were incubated for 36 hours before they were collected and lysed with passive lysis buffer to extract total protein. Protein quantification was conducted using BCA protein assay kit (Pierce, Waltham, MA, USA) following the manufacturer’s instructions. Protein samples of the untreated group, miR-19 mimic-transfected group, and miR-NC-transfected group were subjected to SDS-PAGE and transferred onto polyvinylidene difluoride membranes. Sequentially, the membranes were incubated with rabbit anti-SIRT1, rabbit anti-GAPDH, and goat anti-rabbit secondary antibodies (Thermo Fisher Scientific) and visualized via enhanced chemiluminescence system. GAPDH was used as an internal reference to normalize the quantity of the protein.
CCK8 assay
For cell proliferation assay, 1×104 transfected cells were plated into 96-well plates and cultured for 6 hours in an incubator. Then, 10 μL cell counting kit-8 (CCK8) reagent (Dojindo Molecular Technologies, Kumamoto, Japan) was cautiously added to each well. The absorbance at 450 nm was determined with a microplate reader (Molecular Devices, San Jose, CA, USA).
Scratch assay
Briefly, 5×105 cells were seeded into 12-well plates. After reaching 90% confluency, the cells at the bottom of the well were scratched using a 10 μL sterilized pipette. The cells were washed three times with PBS and cultured with fresh medium containing 2.5% low serum in an incubator at 37°C with 5% CO2. Cell mobility was analyzed by comparing the width of scratches after 24 hours with the initial width using a microscope (Olympus Corporation, Tokyo, Japan).
Transwell assay
Transwell chamber (Corning Costar Corporation, Corning, NY, USA) experiments were performed to analyze the migration and invasion abilities of transfected cells. For this assay, 1×105 transfected cells were added to the upper chamber of the transwell and 600 μL of medium containing 10% FBS was added to the lower chamber. After incubation, the cells were scraped, stained, and observed under an optical microscope (Phenix Optical Instrument Group Company, Jiangxi, China). The invasion ability was determined by calculating the number of invasive cells in four random fields.
Flow cytometry assay
After 48 hours of transfection, A549 cells were trypsinized and harvested for detecting and analyzing the cell cycle using BD FACSCalibur flow cytometry (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). During the experiment, the cells were washed two times with PBS and fixed with 70% ethanol at 4°C. After 24 hours, the fixative was discarded and the cells were washed and resuspended with PBS to reach a concentration of 1×104 cells. Then, appropriate amount of stock solution was added and the cells were incubated at 37°C in a water bath for 30 minutes. After RNA removal, the cells were stained with 50 μg/mL propidium iodide solution. The detection was performed on a computer, and the experiment was repeated three times.
Immunohistochemical assay
The tissue samples were made into 4 μm-thick sections. The sections were deparaffinized and rehydrated, and the antigens were unmasked by heat-induced epitope retrieval for 1 hour. Then, these samples were incubated with rabbit anti-SIRT1, rabbit anti-GAPDH, and goat anti-rabbit secondary antibodies (Thermo Fisher Scientific) to observe the CBX7 expression level.
A549 cells transfected with miR-19 mimics or miR-NC were subcutaneously injected into nude mice. The blank control group received the same dose of physiological saline. From the third day, tumor volume was determined every three days. After 21 days, the lung tumor tissues and normal lung tissues were surgically resected. The formalin-fixed and paraffin-embedded tissues were analyzed to detect the expression level of proliferating cell nuclear antigen (PCNA) (PCNA staining test) and the change of apoptotic cells (TUNEL assay).
Statistical analysis
All the experiments were performed three times, and the results were reported as mean ± SD. Statistical analyses were performed using SPSS software version 17.0 (SPSS Inc., Chicago, IL, USA). Student’s t-test was applied to evaluate the differences between two samples, and Fisher’s exact test or chi-squared test was used to assess the differences in proportion. Differences were considered to be significant at P<0.05.
Results
miR-19 expression in tissues and cell lines
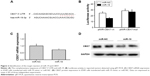
To examine the impact of lung cancer on miR-19 expression, we detected miR-19 expression in NSCLC tissue (n=15) and non-tumor tissue samples (n=15) using qRT-PCR. The results demonstrated that miR-19 expression was significantly increased in cancer tissues compared with non-tumor samples (Figure 1A). Furthermore, the expression level of miR-19 in tumor tissues with metastasis and diameter greater than 5 cm was significantly higher than those in tumor tissues without metastasis and with diameter less than 5 cm (Figure 1B and D). Meanwhile, lung cancer cell lines including NCL-H460, NCL-H1975, 95D, HCL-H358, A549, and NCL-H1299 and control cell line BEAS-2B were also analyzed, and the results showed that the expression level of miR-19 in lung cancer cells was significantly upregulated (Figure 1C). The statistical analysis showed that overexpression of miR-19 is associated with lymph node metastasis and clinical stage of NSCLC (Table 1).
  | Table 1 Correlation between miR-19 expression in tumor tissues and clinicopathological characteristics of NSCLC patients |
Identification of the target reaction of miR-19 and CBX7
CBX7 was identified as the target gene of miR-19 as per verification and computational prediction through miRNA target gene database (Figure 2A). The luciferase reporter vectors pMIR-CBX7-wt and pMIR-CBX7-mut, which contained the wild-type and mutant potential binding sequence of 3′-UTR of CBX7, were established and co-transfected with miR-19 mimic or miR-NC into A549 cells to further study the target reaction of miR-19 and CBX7.
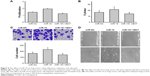
The results of the luciferase reporter assay showed that the relative luciferase activity in pMIR-CBX7-wt+ miR-19 mimic group was significantly reduced compared with the pMIR-CBX7-wt+ miR-NC group. However, there was no obvious change in luciferase activity between the pMIR-CBX7-mut+ miR-19 mimic group and the pMIR-CBX7-mut+ miR-NC group (Figure 2B). Thus, the results showed that miR-19 could inhibit CBX7 expression via binding to the 3′-UTR of CBX7 (Figure 2B). Moreover, qRT-PCR assay indicated that the expression of CBX7 mRNA in miR-19 mimic-transfected group was significantly reduced compared with the control group (Figure 2C) and clarified the phenomenon that miR-19 could inhibit the expression of CBX7 mRNA. Furthermore, we adopted Western blot assay to detect the effect of miR-19 on the expression of CBX7 protein, and the results showed that CBX7 protein expression in the miR-19 mimic-transfected group was significantly decreased (Figure 2D). Thus, the experimental results indicated that miR-19 could inhibit CBX7 expression at the protein level and promote degradation of CBX7 mRNA.
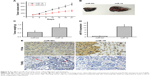
Immunohistochemical assay was performed to detect the expression level of CBX7 in the lung tumor tissue and adjacent normal tissue. The results showed that the expression level of CBX7 in the lung tumor tissue was significantly lower than that in the non-tumor tissue (Figure 3A). Moreover, qRT-PCR revealed that CBX7 mRNA expression was significantly decreased in the lung tumor tissue compared with the non-tumor tissue (Figure 3B). A curve of CBX7 mRNA expression vs miR-19 expression was generated, and a reverse correlation was found (Figure 3C). Further experiments to examine the mRNA expression level of CBX7 in lung cancer cell lines were also conducted. As Figure 3D shows, CBX7 mRNA expression in each lung cancer cell line was significantly downregulated. Thus, the results indicated that CBX7 is the target gene of miR-19, and miR-19 is involved in the development and progression of lung cancer via inhibiting CBX7 expression.
miR-19 affects the proliferation, migration, and cell cycle of lung cancer cells by inhibiting CBX7 expression
To further confirm that the influence of miR-19 on lung cancer cells was dependent on the inhibition of CBX7 expression, we studied the cell proliferation, migration, and invasion capacities using CCK8 assay, transwell assay, and scratch assay, respectively. miR-19 mimic and miR-NC were transfected into A549 cells. Besides, miR-19 and CBX7 were also transfected together into those cells. The results of CCK8 assay exhibited that the proliferation of cells in miR-19 mimic-transfected group was significantly higher than that in the control group, and was significantly increased than that of miR-19 mimic+ CBX7-transfected group (Figure 4A). Flow cytometry assay was carried out to detect the cell cycle phase of cells in each group. Data revealed that the ratio of A549 cells at S phase in miR-19 mimic-transfected group was significantly higher than that in control group, while the ratio in miR-19 mimic+ CBX7-transfected group was significantly lower than that in miR-19 mimic group (Figure 4B). Transwell experiments elucidated that the cell migration in miR-19 mimic-transfected group was significantly higher than that in control group, whereas the cell migration in miR-19 mimic+ CBX7 group was significantly lower than that in miR-19 mimic-transfected group (Figure 4C). Consistently, scratch test also yielded similar experimental results (Figure 4D). Thus, the results showed that miR-19 affects the proliferation, migration, and cell cycle of lung cancer cells by inhibiting CBX7 expression.
A549 cells transfected with miR-19 mimic or miR-NC were subcutaneously injected into nude mice. After 21 days, tumor volume, tumor weight, and miR-19 expression in miR-19 mimic-transfected group were found to be significantly higher than those in miR-NC group (Figure 5A–D). PCNA staining test showed that miR-19-treated tumor cells showed significantly increased proliferation, whereas TUNEL assay showed that cell apoptosis was remarkably reduced compared with the control group (Figure 5E). Thus, the results confirmed that miR-19 promotes lung cancer cell proliferation and inhibits cell apoptosis.
Discussion
Lung cancer is one of the most harmful malignancies, and its morbidity and mortality rates show a global upward trend. With the development of molecular biology, targeted therapy has become the focus of research on individualized treatment of lung cancer. In recent decades, extensive researches have revealed that miRNAs play critical roles in tumor cell proliferation, differentiation, apoptosis, invasion, and metastasis by targeting the related protein molecules. AGR2 is a well-studied secreted protein that contributes to cell proliferation and migration, and miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in NSCLC.25 YAP is associated with the development of multiple tumors, and miR-361 targets 3′-UTR of YAP mRNA to inhibit the proliferation of lung cancer cells.26 miR-455-3p has been reported to act as a prognostic factor and to regulate the biological processes of NSCLC via targeting homeobox B5.27 Additionally, double knockdown of miR-1254 and SFRP1 promoted cell proliferation of lung cancer, demonstrating that miR-1254 promotes lung cancer cell proliferation by targeting SFRP1.28 Thus, many miRNAs play critical roles in regulating lung cancer initiation and progression.
miR-19 is a critical carcinogenic miRNA that participates in important biological processes of human malignancies, such as gastric cancer and osteosarcoma.29,30 Previous researches have showed that miR-19 is involved in the metastasis and development of NSCLC, which indicates that overexpression of miR-19 might be a negative prognostic factor for NSCLC patients.31 Moreover, a study showed that underexpression of miR-19 inhibited the ability of tumorsphere formation, suppressing lung cancer stem cells.32 CBX7, belonging to the PRC1 class of proteins, plays a key role in the development and progression of lung cancer.33 However, the correlation between miR-19 and CBX7 has not been studied so far.
In the present study, for the first time, we evaluated miR-19 expression in NSCLC tissues and cell lines using qRT-PCR. An overexpression of miR-19 was observed in NSCLC tissues compared with non-tumor samples. Moreover, miR-19 expression was upregulated in tumor tissues with metastasis and diameter greater than 5 cm compared with the NSCLC tissues without metastasis and with tumor diameter less than 5 cm. In addition, we also found that miR-19 expression levels were significantly higher in NSCLC tissues and its expression was associated with lymph node metastasis and clinical stage. Similar results were found when miR-19 expression in lung cancer cell lines was compared with normal cell line. CBX7 was identified as a candidate target of miR-19 in lung cancer using the miRNA target expression database and luciferase reporter assay. By transfecting the miR-19 mimic into A549 lung cancer cells, we found that the expression level of CBX7 protein and mRNA was remarkably reduced. Thus, the expression of miR-19 was found to be inversely correlated with the expression of CBX7 in lung cancer. Immunohistochemical assay showed that the expression level of CBX7 in lung tumor tissues was significantly lower than that in non-tumor tissues. Furthermore, we investigated CBX7 mRNA expression in NSCLC tissues and cell lines. CBX7 mRNA expression level in tumor samples and cancer cell lines was obviously lower than that in normal ones. Additionally, the overexpression of miR-19 could significantly enhance lung cancer cell proliferation and migration. Clearly, miR-19 inhibits CBX7 expression and is involved in the development and progression of lung cancer.
Conclusion
The present study confirmed that CBX7 is a functional target of miR-19 which promotes lung cancer cell proliferation via inhibiting CBX7 expression. The increased expression of miR-21 was strongly associated with lymph node metastasis. The identification of interaction between miR-19 and CBX7 in this research facilitated our understanding of molecular mechanisms of tumor genesis and may help in development of potential therapeutic targets for the treatment of lung cancer.
Author contributions
XP conceived the study and designed the experiments. XP and LG contributed to the data extraction, performed the analysis, and interpreted the results. BG wrote the first draft. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Manchado E, Weissmueller S, Morris JP, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534(7609):647–651. | ||
Wang H, Wu S, Zhao L, et al. Clinical use of microRNAs as potential non-invasive biomarkers for detecting non-small cell lung cancer: a meta-analysis. Respirology. 2015;20(1):56–65. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Nishiyama A, Katakami N, Yoshioka H, et al. Retrospective efficacy and safety analyses of erlotinib, pemetrexed, and docetaxel in EGFR-mutation-negative patients with previously treated advanced non-squamous non-small-cell lung cancer. Lung Cancer. 2015;89(3):301–305. | ||
Sun D, Li X, Ma M, et al. The predictive value and potential mechanisms of miRNA-328 and miRNA-378 for brain metastases in operable and advanced non-small-cell lung cancer. Jpn J Clin Oncol. 2015;45(5):464–473. | ||
Wang P, Yang D, Zhang H, et al. Early detection of lung cancer in serum by a panel of microRNA biomarkers. Clin Lung Cancer. 2015;16(4):313–319. | ||
Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. | ||
Hannafon BN, Sebastiani P, de Las Morenas A, Lu J, Rosenberg CL. Expression of microRNA and their gene targets are dysregulated in preinvasive breast cancer. Breast Cancer Res. 2011;13(2):R24. | ||
Zhang X, Yu H, Lou JR, et al. MicroRNA-19 (miR-19) regulates tissue factor expression in breast cancer cells. J Biol Chem. 2011;286(2):1429–1435. | ||
Bruce JP, Hui AB, Shi W, et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget. 2015;6(6):4537–4550. | ||
Ben-Hamo R, Efroni S. MicroRNA regulation of molecular pathways as a generic mechanism and as a core disease phenotype. Oncotarget. 2015;6(3):1594–1604. | ||
Mirzaei H, Masoudifar A, Sahebkar A, et al. MicroRNA: A novel target of curcumin in cancer therapy. J Cell Physiol. 2018;233(4):3004–3015. | ||
Yan L, Zhao W, Yu H, et al. A comprehensive meta-analysis of microRNAs for predicting colorectal cancer. Medicine. 2016;95(9):e2738. | ||
Jili S, Eryong L, Lijuan L, Chao Z. RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem Funct. 2016;34(8):597–605. | ||
Yang B, Jia L, Guo Q, et al. MiR-564 functions as a tumor suppressor in human lung cancer by targeting ZIC3. Biochem Biophys Res Commun. 2015;467(4):690–696. | ||
Tian L, Shan W, Zhang Y, et al. Erratum to: Up-Regulation of miR-21 Expression Predicate Advanced Clinicopathological Features and Poor Prognosis in Patients with Non-Small Cell Lung Cancer. Pathol Oncol Res. 2016;22(2):439. | ||
Wang W, Zhang A, Hao Y, Wang G, Jia Z. The emerging role of miR-19 in glioma. J Cell Mol Med. 2018;22:4611–4616. | ||
Wu Q, Yang Z, An Y, et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144. | ||
Olive V, Bennett MJ, Walker JC, et al. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23(24):2839–2849. | ||
Liu C, Wu H, Li Y, et al. SALL4 suppresses PTEN expression to promote glioma cell proliferation via PI3K/AKT signaling pathway. J Neurooncol. 2017;135(2):263–272. | ||
Zhang XW, Zhang L, Qin W, et al. Oncogenic role of the chromobox protein CBX7 in gastric cancer. J Exp Clin Cancer Res. 2010;29:114. | ||
Shinjo K, Yamashita Y, Yamamoto E, et al. Expression of chromobox homolog 7 (CBX7) is associated with poor prognosis in ovarian clear cell adenocarcinoma via TRAIL-induced apoptotic pathway regulation. Int J Cancer. 2014;135(2):308–318. | ||
Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018;11(1):17. | ||
Pallante P, Terracciano L, Carafa V, et al. The loss of the CBX7 gene expression represents an adverse prognostic marker for survival of colon carcinoma patients. Eur J Cancer. 2010;46(12):2304–2313. | ||
Xue X, Fei X, Hou W, et al. miR-342-3p suppresses cell proliferation and migration by targeting AGR2 in non-small cell lung cancer. Cancer Lett. 2018;412:170–178. | ||
Zhang S, Liu Z, Wu L, Wang Y. MiR-361 targets Yes-associated protein (YAP) mRNA to suppress cell proliferation in lung cancer. Biochem Biophys Res Commun. 2017;492(3):468–473. | ||
Gao X, Zhao H, Diao C, et al. miR-455-3p serves as prognostic factor and regulates the proliferation and migration of non-small cell lung cancer through targeting HOXB5. Biochem Biophys Res Commun. 2018;495(1):1074–1080. | ||
Li H, Yang T, Shang D, Sun Z. miR-1254 promotes lung cancer cell proliferation by targeting SFRP1. Biomed Pharmacother. 2017;92:913–918. | ||
Wang F, Li T, Zhang B, et al. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun. 2013;434(3):688–694. | ||
Sun Z, Liu Q, Hong H, Zhang H, Zhang T. miR-19 promotes osteosarcoma progression by targeting SOCS6. Biochem Biophys Res Commun. 2018;495(1):1363–1369. | ||
Li J, Yang S, Yan W, et al. MicroRNA-19 triggers epithelial-mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95(9):1056–1070. | ||
Zhu J, Wang S, Chen Y, et al. miR-19 targeting of GSK3β mediates sulforaphane suppression of lung cancer stem cells. J Nutr Biochem. 2017;44:80–91. | ||
Forzati F, Federico A, Pallante P, et al. CBX7 is a tumor suppressor in mice and humans. J Clin Invest. 2012;122(2):612–623. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.