Back to Journals » Cancer Management and Research » Volume 14
MIR31HG Expression Predicts Poor Prognosis and Promotes Colorectal Cancer Progression
Authors Wang J, Liu B, Cao J, Zhao L, Wang G
Received 30 November 2021
Accepted for publication 28 May 2022
Published 16 June 2022 Volume 2022:14 Pages 1973—1986
DOI https://doi.org/10.2147/CMAR.S351928
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Ahmet Emre Eşkazan
Jianlong Wang,1,2 Bin Liu,3 Jiewei Cao,3 Lianmei Zhao,4 Guiying Wang2,5
1Department of General surgery, the Second Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China; 2Department of General Surgery, the Fourth Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China; 3Central Laboratory, the Second Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China; 4Department of Research Center, the Fourth Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China; 5Department of General Surgery, the Third Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China
Correspondence: Guiying Wang, Department of General Surgery, the Third Hospital of Hebei Medical University, Shijiazhuang, People’s Republic of China, Email [email protected]
Purpose: Long noncoding RNAs (lncRNAs) are correlated with cancer pathogenesis and prognosis. Many studies have shown that aberrant expression of MIR31HG is implicated in the cancer progression and patient prognosis. However, the biological function and predictive value of MIR31HG in colorectal cancer is unclear.
Methods: The correlation between MIR31HG expression and clinicopathological characteristics of colorectal cancer patients was analyzed by collating the information from The Cancer Genome Atlas (TCGA) database. Kaplan–Meier analysis, univariable and multivariable Cox regression analysis were performed to evaluate the prognostic value of MIR31HG. Gene set enrichment analysis (GSEA) was conducted to identify the potential carcinogenic mechanisms implicated in MIR31HG. Moreover, MIR31HG was knocked down using siRNA in colorectal cancer cells, and cell migration, invasion, growth and colony formation assays were performed. The expression of MIR31HG influenced gene markers was quantified by qRT-PCR in MIR31HG-silenced colorectal cancer cells.
Results: In TCGA database, we found that MIR31HG was elevated in colorectal cancer patients. The patients with high MIR31HG expression had poor overall survival and disease-specific survival. Univariable and multivariable analyses showed that MIR31HG expression was an independent prognostic predictor in colorectal cancer patients. GSEA revealed that MIR31HG mainly modulated focal adhesion, extracellular matrix organization, integrin cell surface interactions and focal adhesion-PI3K-Akt-mTOR-signaling pathway. Besides, MIR31HG knockdown significantly impaired colorectal cancer cell migration, invasion, growth and colony formation. Further qRT-PCR data confirmed that alteration of MIR31HG expression notably affected the tumorigenesis-related key gene expression in the cells.
Conclusion: Our findings provide evidence that MIR31HG is a key factor in maintaining the malignant phenotype of colorectal cancer and may act as an independent predictor for patients with colorectal cancer.
Keywords: colorectal cancer, lncRNA, MIR31HG, prognosis
Introduction
Colorectal cancer is the second and third most common cancer among women and men, respectively. Globally, more than 1.85 million patients are diagnosed with colorectal cancer annually.1 Despite various treatment strategies including drug therapy, radiotherapy, and surgical resection have made great progress in recent years,2,3 the high incidence of recurrence and distant metastasis still affects a large percentage of patients. Due to the complexity of tumorigenesis, the treatment of these patients remains challenging.1,4 Therefore, it is necessary to identify more effective targets for the diagnosis, treatment and prediction of colorectal cancer.
Long non-coding RNAs (lncRNAs) are a class of RNA transcripts over 200 nucleotides and are not capable of coding for proteins.5 Many studies have shown that lncRNAs play critical roles in a variety of biological processes, such as cell proliferation, apoptosis, migration, and invasion.5–7 The expression of lncRNAs is frequently dysregulated in cancers, and some of them are correlated with cancer pathogenesis and prognosis.8–10 A growing number of mechanistic studies have revealed that lncRNAs can interact with certain functional proteins to modify gene expression at the transcriptional or post-transcriptional level.10,11 Despite many new findings, the understandings of the expression profiles and biological functions of lncRNAs in cancers remain limited. Recently, some studies have been conducted to investigate the prognostic relationship between LncRNA microRNA-31 host gene (MIR31HG, also known as LOC554202) and cancers. MIR31HG is mainly located in the cytoplasm and is involved in regulating the target gene expression in cancer cells.12 Aberrant expression of MIR31HG has been found to be significantly correlated with survival time in various cancers, including bladder cancer,13 esophageal squamous cell carcinoma,14 pancreatic ductal adenocarcinoma,15 and colorectal cancer,16 indicating that MIR31HG not only has oncogene functions, but also is a promising biomarker for molecular pathology diagnosis and prognosis prediction in cancer patients. A recent study showed that MIR31HG expression was lower in colorectal cancer tissues than in adjacent noncancerous tissues,17 and the colorectal cancer patients with low expression of MIR31HG implicated poor overall survival and disease-free survival.18,19 However, another study indicated that MIR31HG expression was significantly higher in colorectal cancer than in adjacent tissues.20 These finding suggested that the role of MIR31HG in cancers was still controversial. Therefore, further studies are necessary to investigate the functional role and predictive value of MIR31HG in colorectal cancer.
In our study, we evaluated the prognostic implication of MIR31HG gene expression in colorectal cancer by bioinformatically analyzing clinical features and survival information in the Cancer Genome Atlas (TCGA) database. Moreover, we conducted in vitro experiments to examine the effects of MIR31HG on the proliferation and invasion of colorectal cancer cells and the associated crucial signaling pathways. Our results suggest that MIR31HG plays a crucial role in cancer progression and it is valuable for predicting the prognosis of colorectal cancer patients.
Materials and Methods
Gene Expression and Clinical Features
The relevant data were downloaded from TCGA database. A total of 478 colorectal cancer patients with the corresponding clinical and general information were collected in this study.
Sample Collection
Colorectal cancer and adjacent tissues were collected from patients stored in liquid nitrogen and stored at −80 °C until RNA extraction. The study was approved by the Ethics Committee of the Second Hospital of Hebei Medical University (Hebei, China) and conducted in accordance with the Declaration of Helsinki.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, USA) as described previously,21 and RNA was reversely transcribed into cDNA using Transcription First Strand cDNA synthesis kit (Roche, Basel, Switzerland). qRT-PCR analyses were quantified with SYBR PrimeScript RT-PCR kit (Takara, Dalian, China). The relative expression of each specific gene was normalized against β-actin level. qRT-PCR primer sequences used in the study were as follows: MIR31HG forward primer, 5’-CAGTTTGGTGGAAGATAA-3’; MIR31HG reverse primer, 5’-TTCTCCTTAATGTCACGCACGATT-3’; TPBG forward primer, 5’-TGGAACTGATCCTGAACCACA-3’, TPBG reverse primer, 5’-CGCGGCAGGTAAAGGAAGT-3’; FYN forward primer, 5’-ATGGGCTGTGTGCAATGTAAG-3’, FYN reverse primer, 5’-GAAGCTGGGGTAGTGCTGAG-3’; ETV1 forward primer, 5’-GGCCCCAGGCAGTTTTATGAT-3’, ETV1 reverse primer, 5’-GATCCTCGCCGTTGGTATGT-3’; TRERF1 forward primer, 5’-CCCTCAAGATACACGGGATGG-3’, TRERF1 reverse primer, 5’-GGTTTCCACGTAGCTGGACAT-3’; β-actin forward primer, 5’-CTCCATCCTGGCCTCGCTGT-3’; β-actin reverse primer, 5’-GCTGTCACCTTCACCGTTCC-3’.
Gene Set Enrichment Analysis (GSEA)
The associations between MIR31HG gene expression and all genes were analyzed using R (v.3.6.3), and then GSEA analysis was performed on the gene expression matrix using the cluster Profiler package in R. P <0.05, and FDR <0.25 were considered as statistically significant.
Cell Culture and Transfection
Human colorectal cancer cell lines HT-29 and SW480 were obtained from the cell bank of the Chinese Academy of Sciences in Shanghai (Shanghai, China). The cells were cultured in DMEM (Gibco, USA) with 10% foetal bovine serum, 100 ng/mL streptomycin and 100 U/mL penicillin. The cells were cultured at 37° C, 5% CO2 in a humidified atmosphere incubator.
Transfection of the cells with siRNAs was performed using Lipofectamine RNAiMAX (Invitrogen, USA). Two different siRNA were synthesized by Genepharma (Shanghai, China) using the following sequences: si-MIR31HG#1: 5’-GCCAUUGAGAAAUCAGGAUTT-3’ (sense) and 5’-AUCCUGAUUUCUCAAUGGCTT-3’ (antisense); si-MIR31HG#2: 5’-GCUUUAAUGGAGCACAAAUTT-3’ (sense) and 5’-AUUUGUGCUCCAUUAAAGCTT-3’ (antisense). Negative control: 5’- UUCUCCGAACGUGUCACGUTT-3’ (sense) and 5’-ACGUGACACGUUCGGAGAATT-3’ (antisense). The efficiency of knockdown was validated by qRT-PCR.
Immunohistochemistry (IHC)
IHC was used to detect protein expression in paraffin-embedded cancer specimens from colorectal cancer patients. After deparaffinization and rehydration, the samples were treated with a 3% hydrogen peroxide to block endogenous peroxidase activity. The sections were incubated with antibodies and then treated with polymer-conjugated horseradish peroxidase. The following antibodies were used: anti-TPBG antibody (Proteintech), anti-FYN antibody (Proteintech), anti-ETV1 antibody (Abcam) and anti-TRERF1 antibody (Proteintech). Standard DAB staining was applied for chromogenic detection of the IHC target proteins.
Western Blot
Proteins were extracted by RIPA buffer. After determination of protein concentrations, the proteins were separated by SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% milk powder and then exposed to primary antibodies against anti-TPBG, anti-FYN, anti-ETV1 and anti-TRERF1 at 4 °C overnight. β-actin (Abcam) was used as an internal control.
Cell Migration and Invasion Assay
Cells were treated and seeded into 24-well Transwell chambers (8 μm pores, Corning, USA) in serum-free medium for 24 h. The bottom wells were filled with complete medium. After incubation for 48 h, the invading cells were fixed and stained with crystal violet. Six random fields were counted and imaged under an inverted microscope. Cell invasion assays were performed using Matrigel-coated chambers with the procedure as described above.
Wound Healing Assay
Cells were treated and cultured in six-well plates until almost completely confluent. Monolayer cells were scratched with the tip of a sterile pipette to create a wound. Photographs of cells were taken at 0 and 48 h after wounding and the width of the wound was measured. The experiment was repeated at least three times.
Cell Growth Assay
To evaluate the growth of the cells, a Cell Counting Kit-8 (CCK-8, Biosharp, China) was performed according to the manufacturer’s instructions. Briefly, cells were treated and plated in 96-well plates. Absorbance of 450 nm was measured using a microplate reader at indicated time. The experiment was independently repeated three times.
Colony Formation Assay
Tumor cells were seeded in 6-well plates (200 cells/well) and treated for 14 days, then the cells were fixed with methanol and stained with 1% crystal violet solution. The number of colonies was counted from three separate plates, and representative photographs were taken.
Statistical Analysis
The analyses were carried out by R version 3.6.3 and corresponding packages. The survival curves were evaluated by the Kaplan–Meier method with the Log rank test applied for comparison. The ROC curve was used to evaluate the diagnostic value of MIR31HG expression. Univariable and multivariable Cox analyses were used to explore independent prognostic factors. The significance of the differences between groups was estimated using a one- or two-way ANOVA tests. Difference was considered statistically significant at P <0.05.
Results
The Expression of MIR31HG in Colorectal Cancer Patients
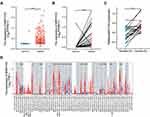
As shown in Table 1, the characteristics of 478 patients with colorectal cancer were acquired from TCGA database. Among the participants, 252 were male (52.7%) and 226 were female (47.3%). To evaluate the expression of MIR31HG in colorectal cancer patients, we compared MIR31HG expression to that in normal colon tissues. The results indicated that the MIR31HG expression was markedly increased in colorectal cancer tissues than in normal tissues (P < 0.001) (Figure 1A). This finding was confirmed in colorectal cancer tissues and paired normal colon tissues (P < 0.001) (Figure 1B). To further validate the data from TCGA, we also measured MIR31HG expression in 22 matched pairs of clinical specimens. The results revealed that MIR31HG was significantly upregulated in cancer tissues compared with normal adjacent tissues (P < 0.001, Figure 1C). Moreover, we also found that MIR31HG was overexpressed in colorectal cancer by analysis of the TIMER database (Figure 1D). Importantly, MIR31HG overexpression can be observed in many human cancers, such as breast cancer, cholangiocarcinoma, esophageal carcinoma, glioblastoma multiforme, and various other cancers (Figure 1D).
 |
Table 1 The Clinicopathological Characteristics of the Colorectal Cancer Patients |
Relationship Between MIR31HG Expression and Clinicopathological Characteristics
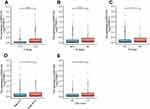
To evaluate the clinical significance of MIR31HG in colorectal cancer patients, we investigated the relationship between MIR31HG expression and clinicopathological characteristics. It showed that high expression of MIR31HG was significantly correlated with T stage (P < 0.001) (Figure 2A), N stage (P < 0.001) (Figure 2B), M stage (P < 0.001) (Figure 2C), pathologic stage (P < 0.05) (Figure 2D) and CEA level (P < 0.05) (Figure 2E), while there was no significant relationship with other clinicopathologic characteristics. Logistic regression indicated that MIR31HG gene expression was a categorical dependent variable related to prognostic poor clinical features (Table 2).
 |
Table 2 Logistic Analysis of the Relationship Between MIR31HG Expression and Clinicopathological Characteristics |
High MIR31HG Expression is Associated with Colorectal Cancer Poor Prognosis
Kaplan–Meier survival analysis was conducted to compare the prognosis of patients dichotomized by MIR31HG expression. The patients with high MIR31HG expression had dramatically worse overall survival (OS, P = 0.033) (Figure 3A) and disease-specific survival (DSS, P = 0.024) (Figure 3B) than patients with low MIR31HG expression. Subgroup analysis by different clinical characteristics showed that high MIR31HG expression was related to poor prognosis in T3-4 stage (P = 0.026), N0-1 stage (P = 0.017), M0 stage (P = 0.007), pathological stage I–II (P = 0.008), no history of colonic polyps (P = 0.003), and more than 65 years old (P = 0.044) (Figure 3C–H). Moreover, similar to the prognostic values of T stage (T3-4 versus T1-2), N stage (N2 versus N0-1), M stage (M1 versus M0), pathologic stage (III–IV versus I–II) and CEA level (>5 versus ≤5), univariable cox regression analysis indicated that high MIR31HG expression was another strong prognostic predictor of poor OS (hazard ratio (HR) = 1.520, P = 0.018) and DSS (HR = 1.899, P < 0.001, Table 3). To further validate the value of MIR31HG expression, multivariable analysis was performed to determine the risk evaluation related to OS and DSS. Most notably, high MIR31HG expression was ascertained as an independent prognostic factor for OS (HR = 1.789, P = 0.010, multivariable analysis) and DSS (HR = 1.859, P = 0.007, multivariable analysis; Table 3). The ROC curve analysis was performed on the MIR31HG expression data, and the area was 0.786, indicating a high diagnostic value of MIR31HG expression in colorectal cancer (Figure 3I). In addition, forest plots of multivariable Cox regression analysis were used to evaluate the prognostic effect of clinical characteristics and risk models in the patients (Figure 4A). Moreover, we combined the expression of MIR31HG and the clinical variables to construct a nomogram to predict the survival probability of the patients at 1, 3, and 5 years (Figure 4B). Taken together, our data reveal that MIR31HG levels can be used as an independent prognostic indicator of clinical outcome in colorectal cancer patients.
 |
Table 3 Univariable and Multivariable Cox Regression Analyses of Clinical Characteristics Correlated with Overall Survival and Disease-Specific Survival in Colorectal Cancer Patients |
GSEA Identifies the MIR31HG-Related Biological Pathways
To explore the biological pathways involved in colorectal cancer, GSEA analysis was conduct in the MIR31HG expression data. Enrichment plots of GSEA showed that the gene sets related to focal adhesion (Figure 5A), extracellular matrix organization (Figure 5B), integrin cell surface interactions (Figure 5C), and focal adhesion-PI3K-Akt-mTOR-signaling pathway (Figure 5D) were involved in patients with high MIR31HG expression, indicating that MIR31HG function may be associated with cancer invasion and metastasis.
MIR31HG Contributes to Colorectal Cancer Progression
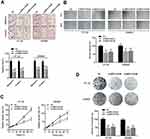
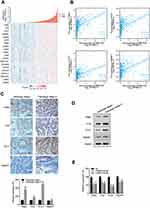
To further understand the cell phenotype associated with MIR31HG in promoting colorectal cancer progression, we knocked down MIR31HG using siRNA in colorectal cancer cells. The transwell and wound healing assays revealed that MIR31HG knockdown resulted in significantly impaired migration and invasion of HT-29 and SW480 cells (Figure 6A and B), which was consistent with the results of Enrichment plots of GSEA. CCK-8 assay showed that colorectal cancer cells with MIR31HG knockdown also grew more slowly than the negative control (Figure 6C). Colony formation assay indicated that MIR31HG knockdown possibly affected the stem cell phenotype (Figure 6D). To predict the function of MIR31HG, we further analyzed the correlation between MIR31HG and the related genes using TCGA database. It showed that high expression of MIR31HG was closely related to the tumorigenesis-related key gene expression of colorectal cancer (Figure 7A), and the MIR31HG expression was positively correlated with that of TPBG (R = 0.480, P < 0.001), FYN (R = 0.480, P < 0.001), ETV1 (R = 0.450, P < 0.001) and TRERF1 (R = 0.460, P < 0.001) (Figure 7B). Moreover, we confirmed the expression of these genes for distinct pathologic stages in colorectal cancer tissues via immunohistochemistry and Western blot analysis (Figure 7C and D). Further qRT-PCR data showed that alteration of MIR31HG expression notably influenced the gene markers participating in tumorigenesis (Figure 7E), indicating that MIR31HG may be a critical regulator in maintaining the malignant phenotype of colorectal cancer.
Discussion
It is revealed that lncRNAs play a crucial role in the pathogenesis of colorectal cancer.22 In recent years, several promising biomarkers of lncRNAs for the diagnosis and prognosis of cancer have been identified. The present study revealed the role of aberrant MIR31HG expression in colorectal cancer progression and patient prognosis. We compared the lncRNA profiles of colorectal cancer and normal colon tissue in the TCGA database and clinical specimens, and we found that MIR31HG showed significantly higher overexpression in colorectal cancer. We further validated this expression pattern in cancer cells and found that high expression of MIR31HG was correlated with colorectal cancer progression. These findings indicated that MIR31HG expression could present an outstanding diagnostic and prospective value for patients with colorectal cancer.
The identification of biomarkers with high sensitivity and specificity is essential for the diagnosis of cancer. With the development of sequencing technologies, lncRNAs have become regulators of a variety of cellular functions through their involvement in epigenetic, transcriptional and post-transcriptional modulation of genes.10,11 An increasing number of studies have shown that the functions of lncRNAs are closely related to cell survival as well as the development of diseases, such as innate immunity,23 pregnancy-induced hypertension,24 and diabetic cardiomyopathy.25 Currently, a large number of studies have identified that MIR31HG is abnormally expressed in a variety of cancers, and overexpression of MIR31HG is considered as a promising prognostic biomarker for cancer patients.14–16 However, the roles and mechanisms of MIR31HG in colorectal cancer are not fully understood, and its prognostic value warrants further exploration.
This study explored the prognostic value of MIR31HG expression in colorectal cancer patients using bioinformatics technology and validated the clinical significance in the patients and cell experiments. Kaplan–Meier survival analysis revealed that overexpression of MIR31HG was closely associated with poor OS and DSS in colorectal cancer patients. Multivariable Cox analysis further supported that high MIR31HG expression was an independent risk factor for OS in the patients, and ROC analysis also proved the diagnostic value of MIR31HG. In addition, we constructed a prognostic nomogram by integrating clinical conditions and gene expression through the TCGA database, enabling clinicians to predict patients’ mortality risk and evaluate the disease to make therapeutic decisions.
To investigate the biological function of MIR31HG, we used GSEA and identified that high expression of MIR31HG was associated with focal adhesion, extracellular matrix organization, integrin cell surface interactions and focal adhesion-PI3K-Akt-mTOR-signaling pathway. In cultured colorectal cancer cells, knockdown of MIR31HG inhibited cell migration, invasion, growth and colony formation. In addition, by analyzing the TCGA database in different MIR31HG expression patients, we found that TPBD, FYN, ETV1 and TRERF1, which were known as characteristic genes involved in cancer cell proliferation and migration,26–29 were significantly suppressed in cancer cells after knockdown of MIR31HG. These results suggested the critical role of MIR31HG in colorectal cancer progression.
Conclusion
In conclusion, our studies found that MIR31HG was upregulated in colorectal cancer and MIR31HG overexpression was closely associated with disease progression. We also identified that MIR31HG played a key role in cancer progression, promoting malignant behaviors by facilitating the invasion and growth of colorectal cancer cells. The effects of MIR31HG on colorectal cancer patients and cells suggested that MIR31HG could be applied as a potential biomarker for the diagnosis and prognosis of colorectal cancer. However, our study is an analysis of the previous published database, and the reliability of the present study requires further molecular mechanistic studies in vitro and in vivo.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 81402510).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA. 2021;325(7):669–685. doi:10.1001/jama.2021.0106
2. Bruni D, Angell HK, Galon J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20(11):662–680. doi:10.1038/s41568-020-0285-7
3. Jung G, Hernández-Illán E, Moreira L, et al. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17(2):111–130. doi:10.1038/s41575-019-0230-y
4. Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21(10):653–667. doi:10.1038/s41577-021-00534-x
5. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
6. Liu SJ, Dang HX, Lim DA, et al. Long noncoding RNAs in cancer metastasis. Nat Rev Cancer. 2021;21(7):446–460. doi:10.1038/s41568-021-00353-1
7. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi:10.1016/j.ccell.2016.03.010
8. Silva-Fisher JM, Dang HX, White NM, et al. Long non-coding RNA RAMS11 promotes metastatic colorectal cancer progression. Nat Commun. 2020;11(1):2156. doi:10.1038/s41467-020-15547-8
9. Zhu Y, Gu L, Lin X, et al. LINC00265 promotes colorectal tumorigenesis via ZMIZ2 and USP7-mediated stabilization of β-catenin. Cell Death Differ. 2020;27(4):1316–1327. doi:10.1038/s41418-019-0417-3
10. Wang FW, Cao CH, Han K, et al. APC-activated long noncoding RNA inhibits colorectal carcinoma pathogenesis through reduction of exosome production. J Clin Invest. 2019;129(2):727–743. doi:10.1172/jci122478
11. Meng N, Chen M, Chen D, et al. Small protein hidden in lncRNA LOC90024 promotes “cancerous” RNA splicing and tumorigenesis. Adv Sci. 2020;7(10):1903233. doi:10.1002/advs.201903233
12. Yan S, Tang Z, Chen K, et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J Exp Clin Cancer Res. 2018;37(1):214. doi:10.1186/s13046-018-0853-9
13. Wu S, Nitschke K, Worst TS, et al. Long noncoding RNA MIR31HG and its splice variants regulate proliferation and migration: prognostic implications for muscle invasive bladder cancer. J Exp Clin Cancer Res. 2020;39(1):288. doi:10.1186/s13046-020-01795-5
14. Chu J, Jia J, Yang L, et al. LncRNA MIR31HG functions as a ceRNA to regulate c-Met function by sponging miR-34a in esophageal squamous cell carcinoma. Biomed Pharmacother. 2020;128:110313. doi:10.1016/j.biopha.2020.110313
15. Yang H, Liu P, Zhang J, et al. Long noncoding RNA MIR31HG exhibits oncogenic property in pancreatic ductal adenocarcinoma and is negatively regulated by miR-193b. Oncogene. 2016;35(28):3647–3657. doi:10.1038/onc.2015.430
16. Eide PW, Eilertsen IA, Sveen A, et al. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int J Cancer. 2019;144(11):2843–2853. doi:10.1002/ijc.31998
17. Ding J, Lu B, Wang J, et al. Long non-coding RNA Loc554202 induces apoptosis in colorectal cancer cells via the caspase cleavage cascades. J Exp Clin Cancer Res. 2015;34(1):100. doi:10.1186/s13046-015-0217-7
18. Rashid NS, Grible JM, Clevenger CV, et al. Breast cancer liver metastasis: current and future treatment approaches. Clin Exp Metastasis. 2021;38(3):263–277. doi:10.1007/s10585-021-10080-4
19. Yang L, Wei H, Xiao HJ. Long non-coding RNA Loc554202 expression as a prognostic factor in patients with colorectal cancer. Eur Rev Med Pharmacol Sci. 2016;20(20):4243–4247.
20. Li Y, Xin S, Wu H, et al. High expression of microRNA-31 and its host gene LOC554202 predict favorable outcomes in patients with colorectal cancer treated with oxaliplatin. Oncol Rep. 2018;40(3):1706–1724. doi:10.3892/or.2018.6571
21. Wang J, Chen X, Hu H, et al. PCAT-1 facilitates breast cancer progression via binding to RACK1 and enhancing oxygen-independent stability of HIF-1α. Mol Ther Nucleic Acids. 2021;24:310–324. doi:10.1016/j.omtn.2021.02.034
22. Han D, Wang M, Ma N, et al. Long noncoding RNAs: novel players in colorectal cancer. Cancer Lett. 2015;361(1):13–21. doi:10.1016/j.canlet.2015.03.002
23. Lai C, Liu L, Liu Q, et al. Long noncoding RNA AVAN promotes antiviral innate immunity by interacting with TRIM25 and enhancing the transcription of FOXO3a. Cell Death Differ. 2021;28(10):2900–2915. doi:10.1038/s41418-021-00791-2
24. Dai C, Zhao C, Xu M, et al. Serum lncRNAs in early pregnancy as potential biomarkers for the prediction of pregnancy-induced hypertension, including preeclampsia. Mol Ther Nucleic Acids. 2021;24:416–425. doi:10.1016/j.omtn.2021.03.010
25. Jakubik D, Fitas A, Eyileten C, et al. MicroRNAs and long non-coding RNAs in the pathophysiological processes of diabetic cardiomyopathy: emerging biomarkers and potential therapeutics. Cardiovasc Diabetol. 2021;20(1):55. doi:10.1186/s12933-021-01245-2
26. Stern PL, Brazzatti J, Sawan S, et al. Understanding and exploiting 5T4 oncofoetal glycoprotein expression. Semin Cancer Biol. 2014;29:13–20. doi:10.1016/j.semcancer.2014.07.004
27. Zhang S, Fan G, Hao Y, et al. Suppression of protein tyrosine phosphatase N23 predisposes to breast tumorigenesis via activation of FYN kinase. Genes Dev. 2017;31(19):1939–1957. doi:10.1101/gad.304261.117
28. Ran L, Murphy D, Sher J, et al. ETV1-positive cells give rise to BRAF(V600E) -mutant gastrointestinal stromal tumors. Cancer Res. 2017;77(14):3758–3765. doi:10.1158/0008-5472.Can-16-3510
29. Gizard F, Robillard R, Barbier O, et al. TReP-132 controls cell proliferation by regulating the expression of the cyclin-dependent kinase inhibitors p21WAF1/Cip1 and p27Kip1. Mol Cell Biol. 2005;25(11):4335–4348. doi:10.1128/mcb.25.11.4335-4348.2005
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.