Back to Journals » OncoTargets and Therapy » Volume 13
MIR155HG Knockdown Inhibited the Progression of Cervical Cancer by Binding SRSF1
Authors Shen L, Li Y, Hu G, Huang Y, Song X, Yu S, Xu X
Received 23 June 2020
Accepted for publication 16 September 2020
Published 23 November 2020 Volume 2020:13 Pages 12043—12054
DOI https://doi.org/10.2147/OTT.S267594
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Ling Shen,1 Yuancheng Li,2 Guiying Hu,3 Yihong Huang,2 Xinli Song,4 Shun Yu,4 Xiaoyuan Xu1
1Department of Obstetrics and Gynecology, The First Affiliated Hospital of Shantou University Medical College, Shantou, Guangdong 515041, People’s Republic of China; 2Department of Gynecological Oncology, The Affiliated Cancer Hospital of Shantou University Medical College, Shantou, Guangdong 515041, People’s Republic of China; 3Department of Gynecology, Maternal and Children Hospital of Guangdong Province, Guangzhou, Guangdong 511400, People’s Republic of China; 4Department of Gynecological Oncology, The Affiliated Cancer Hospital of Shantou University Medical College, Shantou, Guangdong, 515041, People’s Republic of China
Correspondence: Xiaoyuan Xu
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Shantou University Medical College, Shantou, Guangdong 515041, People’s Republic of China
Email [email protected]
Background: As the fourth most common cancer among women worldwide, cervical cancer lead to 311,000 deaths in 2018. Although the treatments have been developed, the survival rate of cervical cancer remains unsatisfactory. In this study, we aimed to identify differentially expressed lncRNAs (DEIncRNAs) between cervical cancer and adjacent normal tissues using bioinformatics analysis, and further to investigate the biological function of the DEIncRNAs in vitro and in vivo.
Methods: The expression profiles from two microarray datasets (GSE6791 and GSE63514) were downloaded from GEO for analysis of DEIncRNAs between cervical cancer and adjacent normal cervical tissues. Among all DEIncRNAs, MIR155HG upregulation was identified and selected for further investigation. The effect of MIR155HG knockdown on proliferation, apoptosis and invasion in SiHa and Hela cells were evaluated. In addition, Western blot, RNA immunoprecipitation (RIP) and cell cycle assays were performed to determine the binding target of MIR155HG. Furthermore, the effect of MIR155HG knockdown on tumor growth in vivo was investigated.
Results: The level of MIR155HG was found to be significantly upregulated in cervical cancer tissue compared with adjacent cervical tissue. Knockdown of MIR155HG notably inhibited the proliferation of SiHa and Hela cells by inducing apoptosis. In addition, MIR155HG knockdown decreased cell invasion. Moreover, tumor growth in xenograft was significantly inhibited by MIR155HG knockdown in vivo. Additionally, SRSF1 was identified as the binding protein of MIR155HG.
Conclusion: Our findings demonstrated that MIR155HG knockdown inhibited the progression of cervical cancer by binding SRSF1, inspiring the usage of MIR155HG as a potential novel therapy target for the treatment of cervical cancer.
Keywords: MIR155HG, cervical cancer, SRSF1
Introduction
Cervical cancer is the fourth most common cancer among women worldwide, affecting 570,000 patients and leading to 311,000 deaths in 2018.1 The major treatments for cervical cancer include surgery, radiotherapy, chemotherapy and target therapy.2 Among these treatment strategies, surgery and radiotherapy are usually considered for patients with localized cancer.2 Concurrent radiochemotherapy is recommended for patients with advanced cervical cancer, and target therapy emerged in recent years are primarily targeting EGFR and COX-2.2 Despite the treatments have been advanced for cervical cancer, the survival rate of cervical cancer is still unsatisfactory.3,4 Therefore, further researches are imperative on understanding the biological mechanisms of cervical cancer progression with the aim of developing novel therapeutic methods.
Long non-coding RNAs (lncRNAs) are important members of the non-coding RNA family with a length of more than 200 nucleotides having no apparent protein coding functions.5 LncRNAs implicate in numerous levels of gene regulation, such as transcriptional, post-transcriptional and epigenetic regulation.6 Thanks to the gene regulation ability of lncRNAs, the associations between dysregulation of lncRNAs and multiple human diseases have been demonstrated, including cancers.6 In addition, the involvement of lncRNAs in multiple cancer processes have been proved such as apoptosis, proliferation, metastasis, and drug resistance.7 The dysregulated expression of lncRNAs was found in a variety of malignancies, such as hepatocellular carcinoma, bladder cancer, breast cancer, gastric cancer, glioma, osteosarcoma, colorectal cancer, and lung cancer.8
Upregulation of lncRNA MIR155 host gene (MIR155HG) was associated with better overall survival (OS) or disease-free survival (DFS) in types of cancers including lung adenocarcinoma, cholangiocarcinoma and skin cutaneous melanoma.9 In contrast, the high expression of MIR155HG was closely related to poorer OS in kidney renal clear cell carcinoma, glioblastoma multiforme, uveal melanoma and brain lower grade glioma.9 However, the role of MIR155HG in cervical cancer remains unclear. In addition, serine/arginine-rich splicing factor 1 (SRSF1) is an RNA binding protein which is directly involved in DNA repair through interaction with non-coding RNA, nascent transcripts, or DNA repair genes.10 It was already reported that SRSF1 played an oncogenic role in cervical cancer.11,12 In this study, we aimed to investigate the role of MIR155HG in cervical cancer both in vitro and in vivo.
Materials and Methods
Bioinformatics Analysis
The expression profiles of GSE6791 and GSE63514 were acquired from Gene Expression Omnibus data base (GEO, https://www.ncbi.nlm.nih.gov/geo/). Gene expression of cervical cancer tissue and site-matched normal tissue was analyzed using R language. LncRNAs with a fold change >2 and adjusted p‑value <0.05 were considered as differentially expressed lncRNAs (DElncRNAs). The levels of MIR155HG in cervical cancer tissue and in adjacent cervical tissue were downloaded from TCGA dataset (http://tcga-data.nci.nih.gov/tcga).
Cell Culture
SiHa and HeLa cells were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). All the cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified atmosphere of 5% CO2 at 37°C. Non-cancerous ectocervical epithelial cell line (Ect1/E6E7) were acquired from ATCC and maintained in Keratinocyte-Serum Free medium (GIBCO, Grand Island, USA) supplemented with 0.05 mg/mL bovine pituitary extract, 0.1 ng/mL human recombinant EGF and 0.4 mM calcium chloride in a humidified incubator with 5% CO2 at 37°C.
Cell Transfection and Lentiviral Infection
Short-hairpin RNAs (shRNAs) against MIR-155HG were constructed in pcDNA3.1 by GenePharma Co., Ltd. (Shanghai, China). The cells were seeded into 6-well plate and cultured overnight before transfection. Next day, the cells were transfected with shRNAs or blank pcDNA3.1 (vector-ctrl, GenScript, Nanjing, China) with Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocols. For lentiviral infection, the cells were seeded into 6-well plate and cultured overnight before infection. SRSF1-overexpressing and control lentivirus were constructed by GenScript (Nanjing, China). After 72 h of infection, the stable infected cells were selected with 5 µg/mL puromycin (Sigma, Saint Louis, MO, USA).
Cell Viability
Cell counting kit-8 (CCK-8) assay (Beyotime, Shanghai, China) was used to determine the cell viability in accordance with the manufacturer’s protocol. The cells (2×103 cells/well) were seeded into 96-well plates and cultured overnight for adherence. After treatments, the cells were cultured with 10 μL CCK-8 solution for 2 h, and the optical density (OD) values at the absorbance of 450 nm was measured using a microplate reader (BioTek, North Brunswick, NJ, USA).
Colony Formation Assay
The cells were seeded into 6 well plates (500 cell/well) and cultured for 2 weeks. After treatments, the cells were fixed in 4% paraformaldehyde for 60 min and stained with 0.005% crystal violet for 15 min. Colonies containing more than 50 cells/colony were counted.
Apoptosis Assay
The annexin V-FITC/propidium iodide (PI) double-staining method was used to quantify apoptosis using Annexin V-FITC Apoptosis Detection Kit (Thermo Fisher Scientific, Waltham, MA, USA). Briefly, the cells were collected and gently washed three times with PBS. After that, the cells of 4 x 105 were resuspended in 200 μL binding buffer followed by incubation with 5 µL of annexin V-FITC and 10 µL of PI following the manufacturer’s instructions. The apoptosis was analyzed by flow cytometry (Dickinson Franklin Lake, NJ, USA).
Transwell Invasion Assay
The 24‐well transwell chambers with 8 μm pores were obtained from Corning (NY, USA). Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was diluted with DMEM F-12 medium and added to the upper chamber (100 μL/well). After that, 1×105 cells resuspended in 100 μL of DMEM F-12 medium were seeded into the upper chamber. The medium (600 μL/well) supplemented with 10% FBS was added to the lower chamber. After incubation for 48 h, the cells that passed through the filter were fixed with 4% paraformaldehyde and stained with 4% crystal violet for 20 min. The invaded cells were observed under a light microscope.
Real-Time qRT-PCR (RT-qPCR)
Total RNAs from the cells were extracted using TRIzol reagent (Invitrogen, Thermo Fisher), followed by transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA, USA). Realtime PCR quantification was performed using EXPRESS One-Step SYBR™ GreenER™ Kit (Thermo Fisher) on a ABI Prism 7500 Sequence Detection System (Perkin Elmer Inc., Waltham, MA, USA). The thermocycling conditions applied were listed as follows: 98°C for 3 min; followed by 40 cycles at 98°C for 30 s, 60°C for 30 s and 72°C for 30 s. The relative expressions of genes were calculated using the comparative 2−ΔΔCt method. ACTIN was used as the inner control. The primers used are described in Table 1.
 |
Table 1 Sequences of Primers |
Western Blotting
Total proteins of the cells were extracted using protein extraction kit (Beyotime). Concentrations of proteins were determined using BCA kit (Beyotime). Then, the proteins were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by transferred onto PVDF membranes (Thermo Fisher). Primary antibodies against SRSF1 (1:2000, Abcam, Cambridge, MA, USA), CDK2 (1:1000, Abcam), CyclinE1 (1:1000, Abcam), β-actin (1:1000, Abcam) were incubated with the membranes at 4°C for 12 h. After washing with TBST for three times, the membranes were then incubated with corresponding HRP-conjugated secondary antibodies (1:5000, Abcam) at room temperature for 2 h. The protein samples were visualized using an ECL detection kit (Merck Millipore, Billerica, MA, USA). The integrated density of each band was normalized to the corresponding β-actin band.
RNA Immunoprecipitation (RIP)
The potential binding relationship between MIR155HG and SRSF1 was explored using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. Then immunoprecipitated RNAs were subjected to RT-qPCR assays to quantify the enrichment of the target RNA.
Cell Cycle Assay
After collected and washed twice in PBS, the cells of 2 × 105 were resuspended in 200 μL PBS. Then, the cell suspensions were fixed in 75% ethanol (1 mL) at 4°C for overnight. After washing two times with PBS, PI/RNase Staining Buffer solution (500 µL, BD Biosciences, Franklin Lakes, NJ, USA) was added to the cell suspensions and incubated for 30 minutes at room temperature. Cell cycle was analyzed using flow cytometry (Dickinson Franklin Lake, NJ, USA).
Xenograft Tumor Model
BALB/c-nu mice (6 weeks of age) were obtained from Vital River (Beijing, China). All animal experiments were performed in accordance with the principles of the NIH Guide for the Care and Use of Laboratory Animals. All protocols were approved by the Ethics Committee for Laboratory Animal Care and Use of the First Affiliated Hospital of Shantou University Medical College. The mice were randomly separated into three groups (n=5): Control, MIR155HG shRNA2 and MIR155HG shRNA2 + SRSF1 OE. After indicated treatment, SiHa cells from each group (3 × 106 cells/mouse) were subcutaneously injected into the right flanks of the mice. The tumor size was measured using a caliper weekly accordingly to the format: length × width2/2. The mice were sacrificed in 4 weeks and the tumors were harvested and weighed.
Statistical Analysis
All experiments of this study were repeated at least three times independently. Statistical analysis was performed with one-way analysis of variance (ANOVA) and Tukey’s test using GraphPad prism version 6.0 (Graphpad Software, La Jolla, CA). Data were presented as mean ± standard deviation of mean (SD). P < 0.05 was considered significant.
Results
MIR155HG Was Upregulated in Cervical Cancer Tissues
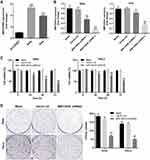
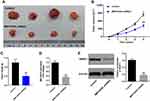
GSE6791 and GSE63514 were firstly downloaded from online dataset of GEO, and the DEIncRNAs were identified and analyzed using R language. As indicated in Figure 1A–D, DElncRNAs between cervical cancer and normal tissue were presented by Volcano Plot. In addition, commonly upregulated or downregulated DElncRNAs in GSE6791 and GSE63514 were illustrated using a Venn diagram. As illustrated in Figure 1E, total 386 DEIncRNAs (270 upregulated and 116 downregulated) were identified in GSE63514 (Figure 1E). Meanwhile, total 241 DEIncRNAs (46 downregulated and 195 upregulated) were identified in GSE6791. Moreover, there were 92 DElncRNAs were commonly dysregulated in these two datasets (85 upregulated and 7 downregulated). Among these commonly dysregulated DElncRNAs, MIR155HG captured our attention due to its influence on OS or DFS in types of cancers.9 Thereby, MIR155HG was selected for further investigation. In addition, the result of TCGA indicated that the median value of MIR155HG in cervical cancer tissue was higher than that in adjacent cervical tissue (Figure 1F). However, no significant difference was observed on overall survival rate between the patients with high and low levels of MIR155HG (Supplementary Figure 1A). Based on these results of bioinformatics analysis and TCGA data, MIR155HG was identified to be unregulated in cervical cancer.
MIR155HG Knockdown Inhibited the Viability of Cervical Cancer Cells
Next, the levels of MIR155HG in cervical cancer cells and in normal cervical epithelial cells were examined by RT-qPCR. The data indicated the level of MIR155HG was upregulated in SiHa and Hela cells compared with that in normal cervical epithelial Ect1/E6E7 cells (Figure 2A). In order to investigate the effect of MIR155HG downregulation in cervical cancer in vitro, SiHa and Hela cells were transfected with MIR155HG shRNA1, MIR155HG shRNA2 or vector control, respectively. The efficiency of shRNA interference was evaluated by RT-qPCR (Figure 2B). Since MIR155HG shRNA2 exhibited best silencing effect, shRNA2 was used in the following experiments. Moreover, CCK-8 assay was used to determine the cell viability at 0, 24, 48, 72 h post-transfection. The results of CCK-8 indicated that MIR155HG knockdown significantly inhibited the viability of SiHa and Hela cells in a time-dependently manner (Figure 2C). Consistently, the result of colony formation assay illustrated that the proliferation of SiHa and Hela cells were repressed by MIR155HG shRNA2 (Figure 2D). Altogether, MIR155HG knockdown significantly inhibited the viability of cervical cancer cells.
MIR155HG Knockdown Increased the Apoptosis and Decreased the Invasion of Cervical Cancer Cells
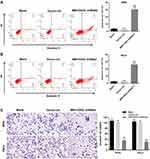
Other than cell viability, the effect of MIR155HG knockdown on cell apoptosis was also detected. As shown in Figure 3A and B, the apoptosis of SiHa and Hela cells were remarkably induced by MIR155HG shRNA2. In addition, the invasion of SiHa and Hela cells were notably repressed by MIR155HG shRNA2 (Figure 3C). Since SiHa cells were more sensitive to MIR155HG shRNA2 compared with Hela, SiHa cells were used for the following experiments. Additionally, to avoid the off-target effect of shRNA knockdown, the inhibitory effect of MIR155HG shRNA1 on viability, apoptosis and invasion were evaluated in SiHa cells (Supplementary Figure 1B–D). The data indicated MIR155HG shRNA1 inhibited cell proliferation and invasion and induced apoptosis as well. All these results indicated that MIR155HG knockdown increased the apoptosis and decreased the invasion of cervical cancer cells.
Serine/Arginine-Rich Splicing Factor 1 (SRSF1) Was the Binding Protein of MIR155HG
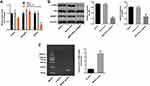
In order to explore the molecular mechanism underlying the anti-tumor effect of MIR155HG knockdown, starBase online database (http://starbase.sysu.edu.cn/) was used to predict the potential binding protein of MIR155HG. There were around 10 RBPs were predicted in starBase online database. Among these RBPs, PTBP1, IGF2BP2 and SRSF1 were predicted to be more likely to interact with MIR155HG base on their binding sites. Next, RT-qPCR was used to verify the binding relationship. The results of RT-qPCR illustrated that the level of SRSF1 was significantly decreased by MIR155HG knockdown, indicating SRSF1 might have a close relationship with MIR155HG (Figure 4A). In consistent with the result of RT-qPCR, the data of Western blot demonstrated that the expression of SRSF1 was downregulated by MIR155HG shRNA2 (Figure 4B). Meanwhile, the protein level of p-Akt was also downregulated by MIR155HG shRNA2, indicating that MIR155HG played its role in cervical cancer cells via affecting PI3K/AKT signaling pathway (Figure 4B). In addition, the potential binding relationship between MIR155HG and SRSF1 was verified by RIP assay (Figure 4C). Thus, SRSF1 was identified as the binding protein of MIR155HG.
MIR155HG Knockdown-Induced Cell Growth Inhibition Was Reversed by SRSF1 Overexpression
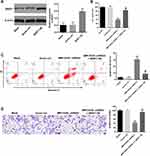
For the purpose of investigating the association between SRSF1 and MIR155HG, SRSF1 overexpressing lentiviruses (SRSF1-OE) was used. Western blot was used to verify the expression of SRSF1 at 72 h after infection. As shown in Figure 5A, the expression of SRSF1 was significantly increased by the infection of SRSF1-OE. Then, the effect of SRSF1-OE on cell viability was detected. The results demonstrated that the cell viability decrease caused by MIR155HG knockdown was rescued by SRSF1 overexpression (Figure 5B). Moreover, since SRSF1 is a splicing factor which regulates apoptosis in cancers.13 The apoptosis assay was performed. The results of apoptosis assay demonstrated that MIR155HG knockdown induced cell apoptosis was attenuated by SRSF1 overexpression (Figure 5C). Consistently, MIR155HG knockdown resulted in the decline of migration in SiHa cells, which was reversed by SRSF1-OE (Figure 5D). All these data illustrated that overexpression of SRSF1 could rescue the effect of MIR155HG knockdown in SiHa cells.
MIR155HG Knockdown-Induced G1 Arrest Was Abrogated by SRSF1 Overexpression
Since SRSF1 acted as a key cell cycle regulator,14 cell cycle analysis was conducted to determine the role of MIR155HG and SRSF1 in cervical cancer. As illustrated in Figure 6A, the percentage of G1 was significantly increased in the group of MIR155HG knockdown compared with the control group. This phenomenon was attenuated in the presence of SRSF1-OE. In addition, the expressions of CDK2 and cyclin E1 were notably decreased by MIR155HG knockdown (Figure 6B). In contrast, MIR155HG knockdown-induced decline in CDK2 and cyclin E1 expression were ameliorated by SRSF1-OE (Figure 6B). Taken together, MIR155HG knockdown exerted anti-tumor action by inducing cell cycle arrest.
MIR155HG Knockdown Inhibited the Growth of Cervical Cancer in vivo
Next, the effect of MIR155HG knockdown on the growth of cervical cancer was investigated in vivo. SiHa cells transfected with MIR155HG shRNA2 or control were injected into the right flanks of nude mice for 4 weeks. As demonstrated in Figure 7A and B, the growth rate of tumor volume markedly decreased in MIR155HG knockdown group. Meanwhile, the average tumor weight of MIR155HG knockdown group was remarkably less than that in control group (Figure 7C). Moreover, the levels of MIR155HG and SRSF1 in the tumor tissue were detected by RT-qPCR and Western blot, respectively. The results indicated that MIR155HG was knocked down by shRNA2 in tumor tissue (Figure 7D). Furthermore, MIR155HG knockdown inhibited the protein expression of SRSF1 (Figure 7E). In addition, the interaction of MIR155HG and SRSF1 in cervical cancer was confirmed in vivo. As shown in Supplementary Figure 2, overexpression of SRSF1 partially reversed the anti-tumor effect of MIR155HG shRNA2 in the xenograft model. Taken together, MIR155HG knockdown notably suppressed the growth of cervical cancer in vivo through inhibiting SRSF1.
Discussion
LncRNAs have emerged as a group of key players in the development of numerous cancers.15 In the present study, we utilized bioinformatics analysis together with cervical cancer tissue sequencing to identify novel valuable lncRNAs for the treatment of cervical cancer. Specifically, MIR155HG was identified and found to be upregulated in cervical cancer tissue. To explore the effect of MIR155HG downregulation on the progression of cervical cancer, MIR155HG was knockdown by shRNA. MIR155HG knockdown inhibited the proliferation, repressed the invasion ability, and increased the apoptosis in cervical cancer cells through its binding protein SRSF1. Moreover, the tumor growth in vivo was also repressed by MIR155HG knockdown. These results suggested that MIR155HG plays a critical role in the progression of cervical cancer. MIR155HG could possibly be used as a potential therapeutic target for the treatment of cervical cancer.
The potentials of MIR155HG as a reliable prognostic and predictive tool for numerous cancers have been proved. In clear cell renal cell carcinoma, MIR155HG together with other 3 lncRNAs (TCL6, PVT1, and HAR1B) were found differentially expressed and correlated significantly with OS of the patients.16 Moreover, in acute myeloid leukemia, the expressions of seven genes, including MIR155HG were associated with OS of the patients.17 Furthermore, MIR155HG overexpression was also found in pancreatic cancer tumor tissue and positively associated with the poor prognosis of the patients. Although MIR155HG has been found involved in these different human cancers, our findings, for the first time, demonstrated the function of MIR155HG in cervical cancer.
MIR155HG mediates it cancer-regulating activity through different molecular mechanisms in types of cancers. According to a study of Colvin et al, MIR155HG played a role in immune response of ovarian cancer by regulating the immune microenvironment.18 Specifically, MIR155HG was upregulated in ovarian cancer-associated fibroblasts (CAFs) and was associated with poor OS of the patients with ovarian cancer. High MIR155HG expression resulted in higher infiltrates of immune cell subsets and enhanced the pro-metastatic role of CAFs.18 However, when MIR155HG played important roles in tumorigenesis and prognosis of kidney renal clear cell carcinoma (KIRC), the underlying mechanism varied. MIR155HG and it’s target gene Cytotoxic T lymphocyte antigen 4 (CTLA-4) were both upregulated in KIRC and associated with OS in patients with KIRC.19 In another previous study, MIR155HG was found overexpressed in non-small cell lung cancer (NSCLC) tumor tissues and was associated with advanced tumor stage and poor prognosis of NSCLC.20 MIR155HG effected NSCLC cells through its two derivatives: miR-155-5p and miR-155-3p. MiR-155-5p and miR-155-3p mediated the anti-NSCLC effect of MIR155HG downregulation in NSCLC cells by negatively regulating the tumor suppressor TP53INP1.20 Moreover, it was recently reported that human MIR155HG was highly expressed in inflamed antigen-presenting cells.21 The highly expressed MIR155HG encoded a 17-amino acid micropeptide which acted as a regulator of antigen presentation and inhibitor of inflammatory diseases.21
Different from these molecular mechanisms, we found that MIR155HG affected the proliferation of cervical cancer cells through the positive correlation with SRSF1. SRSF1 is one of the best studied splicing factors which control splice site recognition in the process of alternative splicing where multiple mRNAs can be generated from the same pre-mRNA.23 Alternative splicing is the major mechanism regulates gene expression in different tissues and disease states through the generation of multiple mRNAs.23 The overexpression of SRSF1 was reported in various type of cancers including lung cancer, kidney cancer, pancreas cancer, breast cancer and so on.23 In cancer cells, SRSF1 can regulate genes involved in cell signaling pathways of cell cycle progression and cell apoptosis.23 In line with previous studies, we demonstrated that SRSF1 overexpression reversed MIR155HG knockdown induced cell cycle arrest through upregulation of CKD2 and cyclin E1. Collectively, the mechanisms underlying cancer-regulation action of MIR155HG were complex. Our findings improved the comprehensive molecular mechanisms of cancer-regulating function of MIR155HG. However, the detailed interaction between MIR155HG, SRSF1 and PI3K/Akt in this study remains to be investigated.
Conclusion
The findings of this study demonstrated that MIR155HG knockdown inhibited the progression of cervical cancer both in vitro and in vivo. The anti-cervical cancer effect of MIR155HG knockdown was realized through it binding protein SRSF1. Therefore, the downregulation of MIR155HG may act as a potential novel therapy against cervical cancer.
Data Sharing Statement
The datasets used in this study are available from the corresponding author on reasonable request.
Ethics Approval
All procedures performed in this study involving animals were approved by the Ethics Committee for Laboratory Animal Care and Use of the First Affiliated Hospital of Shantou University Medical College (ethics approval number SUMC.No20190830b101).
Consent for Publication
Not applicable.
Funding
This study was supported by Natural Science Foundation doctoral program of Guangdong Provincial Science and Technology Department. The role of HIV gp120 in apoptosis of sperm. Grant No.: 2016a030310075.
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Wang L, Zhao Y, Wang Y, Wu X. The role of galectins in cervical cancer biology and progression. Biomed Res Int. 2018;2018:2175927.
3. Zhang JJ, Fan LP. Long non-coding RNA CRNDE enhances cervical cancer progression by suppressing PUMA expression. Biomed Pharmacother. 2019;117:108726. doi:10.1016/j.biopha.2019.108726
4. Zhu H, Zheng T, Yu J, Zhou L, Wang L. LncRNA XIST accelerates cervical cancer progression via upregulating Fus through competitively binding with miR-200a. Biomed Pharmacother. 2018;105:789–797. doi:10.1016/j.biopha.2018.05.053
5. Su K, Zhao Q, Bian A, Wang C, Cai Y, Zhang Y. A novel positive feedback regulation between long noncoding RNA UICC and IL-6/STAT3 signaling promotes cervical cancer progression. Am J Cancer Res. 2018;8(7):1176–1189.
6. Zhu H, Chen X, Hu Y, et al. Long non-coding RNA expression profile in cervical cancer tissues. Oncol Lett. 2017;14(2):1379–1386. doi:10.3892/ol.2017.6319
7. Jiang B, Sun R, Fang S, et al. Lnc-CC3 increases metastasis in cervical cancer by increasing Slug expression. Oncotarget. 2016;7(27):41650–41661. doi:10.18632/oncotarget.9519
8. Cao L, Jin H, Zheng Y, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019;110(3):913–925. doi:10.1111/cas.13921
9. Peng L, Chen Z, Chen Y, Wang X, Tang N. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 2019;8(17):7161–7173. doi:10.1002/cam4.2583
10. Martínez-Terroba E, Ezponda T, Bértolo C, et al. The oncogenic RNA-binding protein SRSF1 regulates LIG1 in non-small cell lung cancer. Lab Invest. 2018;98(12):1562–1574. doi:10.1038/s41374-018-0128-2
11. Kim YJ, Kim BR, Ryu JS, et al. HNRNPA1, a splicing regulator, is an effective target protein for cervical cancer detection: comparison with conventional tumor markers. Int J Gynecol Cancer. 2017;27(2):326–331. doi:10.1097/IGC.0000000000000868
12. Dong M, Dong Z, Zhu X, Zhang Y, Song L. Long non-coding RNA MIR205HG regulates KRT17 and tumor processes in cervical cancer via interaction with SRSF1. Exp Mol Pathol. 2019;111:104322. doi:10.1016/j.yexmp.2019.104322
13. Kędzierska H, Piekiełko-Witkowska A. Splicing factors of SR and hnRNP families as regulators of apoptosis in cancer. Cancer Lett. 2017;396:53–65. doi:10.1016/j.canlet.2017.03.013
14. Zhou X, Li X, Yu L, et al. The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int J Biochem Cell Biol. 2019;113:75–86. doi:10.1016/j.biocel.2019.06.003
15. Wang A, Bao Y, Wu Z, et al. Long noncoding RNA EGFR-AS1 promotes cell growth and metastasis via affecting HuR mediated mRNA stability of EGFR in renal cancer. Cell Death Dis. 2019;10(3):154. doi:10.1038/s41419-019-1331-9
16. Liu H, Ye T, Yang X, et al. A panel of four-lncRNA signature as a potential biomarker for predicting survival in clear cell renal cell carcinoma. J Cancer. 2020;11(14):4274–4283. doi:10.7150/jca.40421
17. Marcucci G, Yan P, Maharry K, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32(6):548–556. doi:10.1200/JCO.2013.50.6337
18. Colvin EK, Howell VM, Mok SC, Samimi G, Vafaee F. Expression of long noncoding RNAs in cancer-associated fibroblasts linked to patient survival in ovarian cancer. Cancer Sci. 2020;111(5):1805–1817. doi:10.1111/cas.14350
19. Song J, Peng J, Zhu C, et al. Identification and validation of two novel prognostic lncRNAs in kidney renal clear cell carcinoma. Cell Physiol Biochem. 2018;48(6):2549–2562. doi:10.1159/000492699
20. Ren XY, Han YD, Lin Q. Long non-coding RNA MIR155HG knockdown suppresses cell proliferation, migration and invasion in NSCLC by upregulating TP53INP1 directly targeted by miR-155-3p and miR-155-5p. Eur Rev Med Pharmacol Sci. 2020;24(9):4822–4835.
21. Niu L, Lou F, Sun Y, et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci Adv. 2020;6(21):eaaz2059. doi:10.1126/sciadv.aaz2059
22. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010;24(11):1073–1074. doi:10.1101/gad.1934910
23. Gonçalves V, Jordan P. Posttranscriptional regulation of splicing factor SRSF1 and its role in cancer cell biology. Biomed Res Int. 2015;2015:287048. doi:10.1155/2015/287048
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.