Back to Journals » OncoTargets and Therapy » Volume 14
miR-944 Suppresses EGF-Induced EMT in Colorectal Cancer Cells by Directly Targeting GATA6
Authors Tang J, Gao W, Liu G, Sheng W, Zhou J , Dong Q, Dong M
Received 30 November 2020
Accepted for publication 14 March 2021
Published 31 March 2021 Volume 2021:14 Pages 2311—2325
DOI https://doi.org/10.2147/OTT.S290567
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
JingTong Tang,1 Wei Gao,1 Gang Liu,1 WeiWei Sheng,1 JianPing Zhou,1 Qi Dong,2 Ming Dong1
1Department of Gastrointestinal Surgery & Hernia and Abdominal Wall Surgery, The First Hospital, China Medical University, Shenyang, 110001, Liaoning, People’s Republic of China; 2Department of General Surgery, The People’s Hospital of China Medical University, Shenyang, People’s Republic of China
Correspondence: JianPing Zhou Fax +86 24-83282886
Email [email protected]
Background: miR-944 belongs to the MicroRNAs family, as shown in our previous study, and is essential in the colorectal cancer (CRC) progression. It is negatively associated with invasion depth and lymph node status. Epithelial-mesenchymal transition (EMT) is essential in tumor invasion and metastasis. However, the relationship between miR-944 and EMT in CRC is unknown and should be further investigated.
Methods: Epithelial–mesenchymal transition (EMT) progression in CRC cell lines was detected with Cell morphology and Western blotting. CRC cell migration and invasion were examined using Transwell assays. Transcriptome and clinical data were obtained from The Cancer Genome Atlas (TCGA) database. The potential pathway of miR-944 and GATA6 were predicted using KEGG analysis. Colocalization was validated using immunofluorescence and Immunohistochemistry. Nuclear and Cytoplasmic Protein Extraction assays were conducted to determine the effects of miR-944 on Wnt/β-catenin signaling.
Results: We found that miR‑944 influences EGF-induced EMT malignant phenotype in vitro. KEGG analyses showed that miR-944 and GATA6 are associated with EMT related pathways, wnt signaling pathways. On the other hand, Western Blot analyses showed that miR-944 can regulate EMT and wnt-β-catenin pathway-related protein, including β-catenin, ZEB1, snail1 via GATA6 regulation. miR-944 also abrogates E-ca after EGF induction. Immunohistochemistry (IHC) and Immunofluorescence (IF) co-expression showed that GATA6 expression is positively associated with β-catenin and ZEB1. GATA6 silencing can reverse EMT malignant phenotype and alterations of related protein induced by miR-944. Quantitative polymerase chain reaction analysis results showed that miR-944 is negatively associated with the UICC stage (P= 0.02), lymph nodes (p=0.04), and liver metastasis (p=0.03). Moreover, patients with high miR-944 expression have better survival (p=0.045). We finally combined miR-944 and GATA6 and found that miR-944/GATA6 ratio could be a novel prognostic biomarker in the TCGA dataset and it is an independent risk prognosis factor (p=0.045).
Conclusion: Our results suggest that miR-944 suppresses the aggressive biological processes by directly repressing GATA6 expression and could be a potential candidate for therapeutic applications in CRC.
Keywords: miR-944, colorectal cancer, GATA6, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) ranks third in cancer incidence rates and is the world’s second cause of cancer-related deaths.1,2 About 25% of CRC patients experience metastasis either at the time of diagnosis or recurrence and most CRC patients with distant metastasis had a poor 5-year survival rate (15%).3 It is therefore important to understand the underlying metastases biological mechanisms of CRC and establish a novel therapeutic target that can help in the CRC malignant process.
The “epithelial-mesenchymal transition” (EMT) (major developmental regulatory program) is essential in promoting tumor invasion and metastasis in colorectal cancer.4 Neoadjuvant therapy can suppress or reverse CRC processes to reduce metastases, recurrence, and resistance.5,6 It is important to find key target playing a significant role in EMT progression for CRC treatment.
GATA6 belongs to the GATA family of transcription factors that binds to the A/TGATAA/G sequence and enhance or suppress downstream gene expression.7 GATA6 is located in the human chromosome (18q11.2) and plays various roles in cancer development.8,9 miR-944 is a conserved miRNA located in the human chromosome (3q28) and it inhibits several cancer cells’ migration and invasion.10,11 Our previous study found that miR-944 inhibits Colorectal cancer (CRC) development by targeting GATA bind protein 6. miR-944 is also negatively associated with CRC patients’ (TNM) stage, invasion depth, and lymph node status.12
This study aimed to determine the miR-944 mechanism on the biological regulation of the EMT process in CRC. We first found that miR-944 suppresses malignant EMT biological behavior in CRC cells and is also negatively associated with CRC patients’ (TNM) stage, lymph node status, and liver distance. CRC patients with low miR-944 expression have poor survival. Furthermore, we confirmed that GATA6 as miR-944 target gene can regulate Wnt–β-catenin pathway, consistent with the KEGG analysis. We finally demonstrated that the low miR-944/GATA6 ratio is associated with poor survival in CRC patients. In conclusion, miR-944 suppresses the biological processes and could be a potential exogenous biological reagent for the CRC therapy.
Materials and Methods
Human Tissue Specimens and Cell Lines
We collected 140 pairs (67 men and 73 women median age of 61.7 years and age range of 29–81 years) of fresh colorectal cancer and their paired adjacent non-tumor colorectal tissues after colorectal cancer surgery at the Gastrointestinal Surgery Department of the First Affiliated Hospital of China Medical University. All the specimens were pathologically diagnosed as colorectal cancer, and the data was classified following the 8th UICC guideline. Informed consent was obtained from some patients, and the remainder were exempt from informed consent as anonymity was assured, as approved by the ethics committee of the First Affiliated Hospital of China Medical University. This study was conducted in accordance with the Declaration of Helsinki. miRNA and mRNA expression data (except a too high or too low expression of miR-944 and GATA6) and the clinicopathological annotations of 285 CRC patients were downloaded from the TCGA portal (http://cancergenome.nih.gov). We analyzed the correlation between GATA6 and β-catenin, β-catenin, and ZEB1 using GEPIA.13
Two CRC cell lines (HCT116 and SW480) were sourced from a cell bank at the Chinese Academy of Sciences (Shanghai, China) and cultured with a recommended growth medium supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) and 100U/m penicillin or streptomycin in a humidified chamber with 5% CO2 at 37°C.
Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis and Gene Set Enrichment Analysis (GSEA)
We classified the samples into high-expression and low-expression miR-944 groups according to the miR-944 expression median levels and conducted KEGG analysis using GSEA. miR-944/GATA6 ratio was also used to conduct a similar analysis.
RNA Isolation and Quantitative Real-Time PCR
We extracted Total RNA from CRC cell lines and colorectal tissue and stored them in liquid nitrogen with Trizol reagent (Takara Bio, Otsu, Japan) according to the instructions. NanoDrop ND-1000 instrument (NanoDrop, USA) was used to measure miR-944, and Hairpin-it microRNA concentration and RT-PCR Quantitation Kit (Shanghai, China) was used to normalize U6 snRNA. We used Cycle Dice Real-Time (Thermal) to measure miRNA expression levels according to instructions with U6 RNA as the internal control. Thermocycling conditions were: 95º C for 3min, then 45 cycles of 95º C for 12s, and 62º C for 45s. The following primers were used: miR‑944 forward primer, 5ʹ‑GCACTCCTAAAATTATTGTACATCG-3ʹ; U6 forward primer, 5ʹ‑CTCGCTTCGGCAGCACA‑3ʹ; miR‑1 reverse primer, 5ʹ‑TATGGTTGTTCACGACTCCTTCAC-3ʹ, U6 reverse primer, 5ʹ‑AAC GCTTCACGAATTTGCGT-3. The relative f miR-944 expression was represented as Fold changes (2 −ΔΔCt) and each experiment was conducted thrice in replicate.
Protein Isolation and Western Blot
RIPA lysis buffer containing 1% PMSF was used to extract the Total sample from two infected CRC cell lines (HCT116 and SW480). We separated protein samples using 10% sodium dodecyl sulfate-polyacrylamide gel and then transferred them on PVDF membranes. The membranes were enclosed with 5% non-fat milk for 2 hours and incubated with a rabbit anti-GATA6 antibody at 1:500 dilution (CST, USA), a rabbit anti-β-catenin antibody 1:1000 dilution (Proteintech, USA), a rabbit anti-ZEB1 (Proteintech, USA) at 1:1000 dilution, a rabbit anti-Lamin B1 (Beyotime, China) at 1:1000 dilution, a mouse anti-GAPDH (Proteintech, USA) at 1:3000 dilution at 4oC for 12 h. The secondary antibodies were used correspondingly for 2 hours. We assessed the Western blot results using the ECL detection kit (Thermo Scientific, Rockford, IL, USA) and each experiment was done thrice.
Immunoprecipitation
Lysis buffer contained 20 mM Tris/HCl (pH7.4), 1.0% NP-40, 150 mM NaCl, 1 mM EDTA, 10 μg/mL leupeptin, and 50 μg/mL PMSF. We used lysis buffer to extract CRC cell protein. Preincubated antibody-beads with magnetic beads (Bio-Rad, Hercules, CA, USA) and 200ul rabbit GATA6-antibody (CST, USA) or IgG (Santa Cruz, Japan) at 1:100 dilution were incubated with extracted CRC protein at 4 °C overnight. We used Western blot the following day to visualize immunoprecipitation sample separated using magnetic beads. We used RIPA lysis to wash immunocomplex.
Immunohistochemistry (IHC)
We deparaffinized Paraffin-embedded PC tissues continuous sections (4um) at 65°C for 2h and then washed them using ethanol and xylene. Antigen repair was done at high pressure for 2.5min for GATA6, β-catenin, and ZEB1. We then incubated sections with hydrogen peroxide (3%) and 10% normal goat serum for 15min and 20 min, respectively. The sections were also incubated with a GATA6 antibody (1:200) (CST, USA), β-catenin antibody (1:200) (Proteintech, USA) and ZEB1 (1:200) (Proteintech, USA) at 4°C overnight. We incubated secondary antibody using streptavidin–peroxidase reagent with sections for 15 min, and then added 3,3′-diaminobenzidine (DAB) for color reactions and visualized at ×20 magnification.
We conducted Immunohistochemistry (IHC) score as previously described.14 GATA6, β-catenin and ZEB1 coloration ranges were used as the standard to assess the staining score defined as 1 (0–25%), 2 (25–50%), 3 (50–75%), 4 (75–100%)[0 (negative), 1 (weak), 2 (moderate) and 3 (strong)]. The two scores were the final score ranging between 0 to 7, and the final score≥ 3 was defined as positive expression.
Transfection
We synthesized miR-944 mimics, miR-944 inhibitor, and GATA6 siRNA and their corresponding negative control (NC) at GenePharma (Shanghai, China) and the sequence were; miR-944 mimics, 5ʹ-AAAUUAUUGUACAUCGGAUGAG-3ʹ, Negative control, 5ʹ-UUCUCCG AACGUGUCACGUTT-3ʹ, miR-944 inhibitor, 5ʹ-CUCAUCCGAUGUACAAUAAUUU-3ʹ, MircoRNA inhibitor N.C, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ. We used Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) to perform transfection according to the manufacturer’s instructions. The transfection efficiency had been tested in our previous study.
EMT Construction
We incubated miR-944 mimics, miR-944 inhibitor, and miR-944 inhibitor + GATA6 siRNA, and their corresponding scramble infected two CRC cell lines (HCT116 and SW480 cells) with 50 ng/mL EGF (Peprotech, RockyHill, NJ, USA) twice for 48 hours using 1% BSA (Sigma) as a control. Cells were cultured with the recommended growth media containing 1% FBS to enhance the EGF effect. We observed EMT-like cell morphology (a spindle-shaped and fibroblast-like morphology), EMT-induced cell invasion and migration, and the alteration of the EMT-related protein, verifying the EMT process.
Cell Migration and Invasion Assay
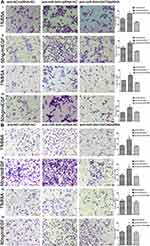
We constructed miR-944 overexpression, HCT116 downregulation, and SW480 cell lines as previously described. We added 8*104infected HCT116/well and 5*105 infected SW480/well in 300ul free medium into the upper chamber and 600 μL medium with 10% fetal bovine serum into the bottom chamber. We coagulated the upper membrane with Matrigel (BD Biosciences) for invasion assays. PC cells stuck in the bottom membrane were stained after 24 hours using cool methanol and dried using 0.1% crystal violet (Sigma) for 30 min. We used a microscope (Nikon Microphot-FX, Tokyo, Japan) to take Cell photographs, and counted the number in five random fields per chamber at ×20 magnification.
Immunofluorescence (IF)
Briefly, we fixed CRC cells plated in 24-well culture plates using 4% paraformaldehyde at room temperature for 30min. The samples were then permeabilized in 1% Triton X-100 and blocked in 5% BSA for 2 h and then incubated with primary antibody [rabbit anti-GATA6 at 1:100 dilution (CST, USA) and mouse anti-β-catenin at 1:100 dilution (Proteintech, USA)] at 4 °C overnight. The secondary antibodies were then incubated with green FITC (Proteintech) and red TRIC (Proteintech). We finally co-stained the cells using DAPI (Sigma, St. Louis, MO, USA) for nuclei visualization and used a fluorescence microscope (Nikon Microphot-FX, Tokyo, Japan) for visualization.
Statistical Analysis
The three independent experiments results were represented as the mean ± standard deviation and we used SPSS 17.0 statistical software (Chicago, IL, USA) to conduct all the statistical analyses. Scientific graphing was plotted using GraphPad Prism (GraphPad Software, USA). Student’s t-test was used to determine the relationship between miR-944 expression, clinicopathological parameters, and cell migration invasion assays. We used Chi-squared, and Cox’s proportional hazards regression model for conducting univariate association analysis between clinicopathological factors and survival to determine independent prognostic factors. Pearson’s correlation coefficient was used to explore the relationship between GATA6 and β-catenin or ZEB1. Protein GATA6, β-catenin, ZEB1, snail1, GATA3, E-cadherin, ZO1 expression results were analyzed using paired t-tests. P<0.05 was considered as a statistically significant difference.
Results
miR‑944 Ectopic Expression Affects EGF-Induced EMT-Like Cell Morphology, and EGF-Induces Cell Malignant Invasion, Migration, and Intracellular Calcium Alteration in Two CRC Cell Lines
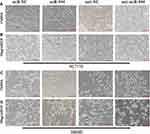
Several studies have found that EGF can induce cell EMT phenotype in various cancers and EMT is essential in cancer metastasis.15,16 We compared the cell morphology between miR-NC and miR-944 groups, anti-NC and anti-944 groups, and the corresponding groups induced by EGF for two days. We found that both HCT116 and SW480 cells exhibited EMT-like cell morphology after EGF introduction and most cells lost their epithelial characteristics and had a spindle-shaped and fibroblast-like morphology. There was no significant cell morphology difference between the miR-NC and miR-944 groups, anti-NC, and anti-944 groups. We found that miR-944 exhibited EMT-like cell morphology and anti-944 had the reverse function after EGF introduction (Figure 1). We compared negative control and transfected groups to determine cancer cell invasion and migration assays. We found HCT 116 and SW480 cell invasion and migration in the miR-944 group with or without EGF treatment (Figure 2). On the contrary, the anti-miR-944 group with or without EGF treatment exhibited different results (Figure 2).
miR-944 Suppresses EMT Activation via the Wnt-β-Catenin Signaling Pathway
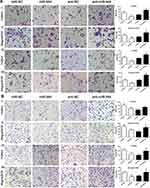
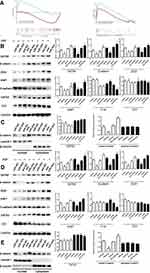
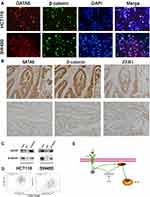
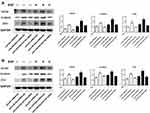
We first conducted KEGG analysis and found that GATA6 was all associated with the Wnt-β-catenin signaling pathway. (Figure 3A, Supplementary Table 1). Wnt-β-catenin signaling pathway is essential in the cancer EMT process.17 Hwang et al18 found that GATA6 directly affects Wnt6, leading to cellular β-catenin accumulation and canonical WNT-β-catenin pathway activation. We identified that miR-944 could bind to the 3ʹUTR of GATA6 in our previous study and hypothesized that miR-944 regulates the wnt-β-catenin signaling pathway. We then extracted protein from transfected HCT116 and SW480 cells to perform Western blot. The results showed that miR‑944 overexpression significantly inhibited GATA6, β-catenin protein and ZEB1 (downstream protein of wnt-β-catenin signaling pathway) and EMT related protein (E-cadherin and snai1l) expression in two CRC cells (Figure 3B and D), and also the β-catenin nuclear translocation (Figure 3C and E). We also found that GATA6 were co-stained in normal HCT116 and SW480 cells in the cytoplasm using IF (Figure 4A) and co-expression relationship between miR-944, GATA6, β-catenin, and ZEB1 using immunohistochemistry. (Figure 4B) Spearman’s rank correlation was used to analyze the results and found consistency with Western Blot results (Table 1). We further identified that GATA6 was structurally binds to β-catenin in two colon cancer cells using IP analysis (Figure 4C). Moreover, the Correlation Analysis showed that GATA6 is positively associated with β-catenin while β-catenin is associated with ZEB1 in GEPIA (Figure 4D).
 |
Table 1 Relationship of GATA6 with miR-944, β-Catenin and ZEB1 |
The intracellular calcium is essential for cellular function regulation, including gene transcription, cell proliferation, invasion, and migration.19,20 We used Fluo-4 and Fluorescence microscope to examine the intracellular calcium.21,22 The Fluorescence intensity in the miR-944 group was weaker than in the miR-NC group. The anti-miR-944 enhanced intracellular calcium in HCT116 and SW480 (Supplementary Figure 1) and EGF-induced EMT was found to be calcium-signal dependent.23 Therefore, miR-944 can regulate CRC EMT phenotype by regulating intracellular calcium translocation.
In summary, miR-944 modulates EMT progress via GATA6-mediated Wnt–β-catenin signaling pathway regulation. (Figure 4E) by
GATA6 Silencing Reverses the Anti-miR‐944 Effects in Both CRC Cell Lines and miR-944 Can Suppress CRC EMT by Binding to GATA6
We conducted a rescue experiment and constructed three co-infected groups (anti-NC and siRNA-NC infected group, anti-miR-944 and siRNA-NC infected group, and anti-miR-944 and GATA6 siRNA infected group) and then compared CRC cell migration and invasion function and measured GATA6, β-catenin, and ZEB1 expression in the three co-infected groups to further identify the relationship between miR-944 and GATA6 and miR-944 mechanism in CRC. GATA6 silencing reversed the inhibition of HCT116 and SW480 cell invasion and migration caused by the miR-944 inhibitor (Figure 5). We also found that GATA6 could reverse GATA6, β-catenin, and ZEB1 expressions induced by miR-944 inhibitor in Two CRC cells (Figure 6).
miR-944 is Associated with Poor Survival and miR-944/GATA6 Ratio is a Prognostic CRC Marker
We used the chi-square test to analyze the relationship between miR-944 expression and clinical parameters in 140 CRC patients. We found that miR-944 is associated with the TNM stage, Lymph node status, and liver metastasis (Table 2). CRC patients were further divided into two groups according to median miR-944 expression level. Kaplan–Meier survival analysis showed that highly expressed miR-944 in CRC patients had a significantly higher overall survival (Figure 7A). We analyzed the relationship between miR-944 expression and clinical parameters in the TCGA dataset and found that miR-944 is negatively associated with the TNM stage, consistent with our results (Figure 7B).
 |
Table 2 Association of miR-944 Expression Levels with Clinical Data |
We further evaluated the combination of miR-944 and GATA6 as a potential CRC prognostic biomarker. We integrated the two markers in a ratio (miR-944/GATA6 ratio) in the TCGA dataset and found that high ratio patients had an early TNM stage and high survival using the R program (4.0.2) (Figure 7C and D). Besides, Univariate and multivariate Cox regression analysis results showed that low miR-944/GATA6 ratio independently causes poor prognosis (Figure 7E and F).
Discussion
EMT (the transformation from benign to invasive carcinoma) are significantly associated with cancer metastasis. However, the EMT process is complex since several genes and signaling pathways are involved in the progress. While several studies have focused on EMT control at molecular levels,24,25 we found that miR-944 downregulation is associated with EMT–related biological processes in CRC (cell adhesion, migration, and invasion). No study has investigated the relationship between miR-944 and EMT and our study provides new information on the EMT mechanism in CRC.
Previous studies have shown that miR-944 suppresses tumors. Flores-Pérez et al26 first reported that miR-944 influences breast cancer cell migration by targeting SIAH1 and PTP4A1. Liu et al27 found that miR-944 as a potential miRNA driver inhibits cell growth in non-small cell lung cancer by targeting EPH7A. While miR-944 has been reported to be oncogenic in cervical cancer,28 subsequent studies have also revealed that it inhibits gastric cancer metastasis by preventing the epithelial-mesenchymal transition via MACC1/Met/AKT signaling.29 We found that miR-944 was negatively associated with CRC patients survival, inhibited CRC EMT phenotype and influenced EMT related protein. Bioinformatics analysis are often used to identify experiments’ results in various cancers.30,31 Meanwhile, our experiments’ results were consistent with the bioinformatics analysis results.
GATA6, a transcription factor gene that is almost amplified or overexpressed in various cancers, is essential in tumor occurrence progress and development.32,33 GATA 6 is involved in the mesoderm and endoderm-derived cell differentiation, including Heart, lungs, and gastrointestinal tract.7 However, the GATA6 mechanism is still unknown. GATA6 overexpression in breast cancer increases epithelial-mesenchymal transition via slug expression upregulation.34 Some studies have also indicated that ectopic expressing microRNAs influence GATA6 expression. miR-196b negatively regulates GATA6 expression in colon cancer and miR-124 induces cell invasion and metastasis by directly targeting GATA6 in cholangiocarcinoma.35,36 We also identified that miR-944 can suppress GATA6 expression and GATA6 inhibition could reverse the effect on EMT phenotype and related protein induced by anti-miR-944 in CRC cells.
Ectopic Wnt activation is common in human cancers, especially in CRC.37 Wnt signaling plays a critical role in development processes, such as influencing stem cell proliferation and differentiation.38 Aberrant activation of canonical Wnt signaling results in β-catenin nuclear translocation found in the dedifferentiated mesenchyme-like tumor cells undergoing an active EMT associated with E-cadherin downregulation.39 It has also been reported that canonical Wnt signaling activation, including TWiST1, ZEB1, and MMP9, amplifies or overexpresses a set of EMT activators.40 Conversely, DKK1, a wnt signaling protein inhibitor, suppresses CRC cell malignant biological behaviors via EMT program inhibition.41 While Transcription factor (GATA6) has been reported to bind to lgr5 promoter regulates LGR5 expression in colorectal tumorigenesis which is downstream of the Wnt-β-catenin signaling pathway.42 The Western Blot and co-expression IHC experiment also indicated that GATA6 is associated with β-catenin and could prompt β-catenin translocation in the nucleus. We hypothesized that the GATA6 expression decrease induced by miR-944 suppresses the EMT process in CRC by mediating the wnt-β-catenin signaling pathway.
Furthermore, miR-944 can regulate the EMT process by mediating Ca2+. The KEGG analysis results showed that miR-944 expression is significantly related to the Calcium signaling pathway. Increasing piece of evidence have shown that intracellular calcium alteration in various cancer cells affects tumor progression, migration, invasion, and metastasis.43,44 Intracellular calcium and intracellular calcium increase in breast cancer abrogate EMT phenotype, resulting in β-catenin nuclear translocation, and this could be the potential canonical Wnt/β-catenin mechanism.23 We found that miR-944 mimics inhibited β-catenin expression in the nucleus. It has also been confirmed that GATA6 as a transcription factor can bind to the calreticulin (CRT) promoter in mice and humans via reporter gene and ChiP analyses.45 Our previous study also found that CRT is positively related to miR-944 and GATA6. CRT regulates the increase of intracellular calcium, Integrinβ1, Fibronectin, and c-Myc expressions induced by EGF in MDCK cells.17 CRT also influences calcium release and influx in integrin-mediated Ca2+ signaling pathway.46,47 Therefore, miR-944-GATA6 regulates CRC EMT phenotype by mediating intracellular calcium.
In conclusion, miR-944-GATA6 suppresses EGF-induced EMT in CRC cells via the wnt/β-catenin signaling pathway which could also be regulated via CRT-mediated change of intracellular calcium. Importantly, the miR-944/GATA6 ratio is a novel biomarker of survival and treatment outcome in CRC patients. However, the relationship between intracellular calcium and EMT, and miR-944-GATA6 mechanism are still unknown and will be further investigated in our future study.
Ethics Approval and Consent
The Ethics Committee of China Medical University authorized all experiments in this study. Written informed consent was obtained from patients.
Consent for Publication
Not applicable.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Scientific Research of Special transformation project of medical science and technology achievements of Liaoning Province, China (Liao Ke Fa, No. 2019–24).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5).
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2). doi:10.3322/caac.21338.
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
4. Cao H, Xu E, Liu H, et al. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol Res Pract. 2015. doi:10.1016/j.prp.2015.05.010
5. Bocca C, Bozzo F, Cannito S, et al. Celecoxib inactivates epithelial-mesenchymal transition stimulated by hypoxia and/or epidermal growth factor in colon cancer cells. Mol Carcinog. 2012;51(10):783–795. doi:10.1002/mc.20846
6. Reka AK, Kuick R, Kurapati H, et al. Identifying inhibitors of epithelial–mesenchymal transition by connectivity map-based systems approach. J Thorac Oncol. 2011;6:1784–1792. doi:10.1097/JTO.0b013e31822adfb0
7. Rojas A, Fernandez MC, Soria B, Martín F, Rojas A. GATA4 and GATA6 control mouse pancreas organogenesis. J Clin Invest. 2012;122(10):3504. doi:10.1172/JCI63240
8. Chen Y, Yang L, Cui T, et al. HOPX is methylated and exerts tumour-suppressive function through Ras-induced senescence in human lung cancer. J Pathol. 2015.
9. Tian F, Li D, Chen J, et al. Aberrant expression of GATA binding protein 6 correlates with poor prognosis and promotes metastasis in cholangiocarcinoma. Eur J Cancer. 2013;49(7):1771–1780. doi:10.1016/j.ejca.2012.12.015
10. Lv L, Wang X, Ma T. microRNA-944 inhibits the malignancy of hepatocellular carcinoma by directly targeting IGF-1R and deactivating the PI3K/Akt signaling pathway. Cancer Manag Res. 2019;11:2531–2543. doi:10.2147/CMAR.S199818
11. Kim YJ, Lee JH, Jin S, et al. Primate-specific miR-944 activates p53-dependent tumor suppression in human colorectal cancers. Cancer Lett. 2018.
12. Tang JT, Zhao J, Sheng W, et al. Ectopic expression of miR-944 impairs colorectal cancer cell proliferation and invasion by targeting GATA binding protein 6. J Cell Mol Med. 2019;23:3483–3494. doi:10.1111/jcmm.14245
13. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi:10.1093/nar/gkx247
14. Sheng W, Dong M, Chen C, et al. Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J off Publ Fed Am Soc Exp Biol. 2017;31(6):2429. doi:10.1096/fj.201601240R
15. Sheng W, Shi X, Lin Y, et al. Musashi2 promotes EGF-induced EMT in pancreatic cancer via ZEB1-ERK/MAPK signaling. J Exp Clin Cancer Res. 2020;39(1).
16. Sheng W, Chen C, Dong M, et al. Calreticulin promotes EGF-induced EMT in pancreatic cancer cells via Integrin/EGFR-ERK/MAPK signaling pathway. Cell Death Dis. 2017;8(10):e3147. doi:10.1038/cddis.2017.547
17. Basu S, Cheriyamundath S, Ben-Ze’Ev A. Cell–cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis. F1000 Res. 2018;7:1488. doi:10.12688/f1000research.15782.1
18. Hwang JTK, Kelly GM. GATA6 and FOXA2 regulate Wnt6 expression during extraembryonic endoderm formation. Stem Cells Dev. 2012;21(17):3220. doi:10.1089/scd.2011.0492
19. Morciano G, Marchi S, Morganti C, et al. Role of mitochondria-associated ER membranes in calcium regulation in cancer-specific settings. Neoplasia. 2018;20(5):510. doi:10.1016/j.neo.2018.03.005
20. Fourbon Y, Guéguinou M, Félix R, et al. Ca2+ protein alpha 1D of CaV1.3 regulates intracellular calcium concentration and migration of colon cancer cells through a non-canonical activity. Sci Rep. 2017;7(1):14199.
21. Chong-Yu Z, Xin-Yuan S, Jian-Ming O, et al. Diethyl citrate and sodium citrate reduce the cytotoxic effects of nanosized hydroxyapatite crystals on mouse vascular smooth muscle cells. Int J Nanomed. 2017;12:8511–8525. doi:10.2147/IJN.S145386
22. Liu L, Yang M, Wang N, et al. New insights of subfertility among transplanted women: immunosuppressive drug FK506 leads to calcium leak and oocyte activation before fertilization. J Cell Biochem. 2017.
23. Davis FM, Azimi I, Faville RA, et al. Induction of epithelial–mesenchymal transition (EMT) in breast cancer cells is calcium signal dependent. Oncogene Basingstoke. 2014;33:2307–2316. doi:10.1038/onc.2013.187
24. Yang Z, Liu S, Zhu M, et al. PS341 inhibits hepatocellular and colorectal cancer cells through the FOXO3/CTNNB1 signaling pathway. Sci Rep. 2016;6:22090.
25. Rokavec M, Öner MG, Li H, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124(4):1853–1867. doi:10.1172/JCI73531
26. Flores-Pérez A, Marchat LA, Rodríguez-Cuevas S, et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer. 2016;16(1):379. doi:10.1186/s12885-016-2470-3
27. Liu M, Zhou K, Cao Y. MicroRNA-944 affects cell growth by targeting EPHA7 in non-small cell lung cancer. Int J Mol Sci. 2016;17(10). doi:10.3390/ijms17101493
28. Liu SS, Chan KKL, Chu DKH, et al. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol Oncol. 2018. doi:10.1002/1878-0261.12383
29. Pan T, Chen W, Yuan X, et al. miR-944 inhibits metastasis of gastric cancer by preventing the epithelial-mesenchymal transition via MACC1/Met/AKT signaling. FEBS Open Bio. 2017;7(7):905–914. doi:10.1002/2211-5463.12215
30. Rahman MR, Islam T, Gov E, et al. Identification of prognostic biomarker signatures and candidate drugs in colorectal cancer: insights from systems biology analysis. Medicina. 2019;55(1):20. doi:10.3390/medicina55010020
31. Islam T, Rahman MR, Shuvo MAH, et al. Drug repositioning and biomarkers in low-grade glioma via bioinformatics approach. Inf Med Unlocked. 2019. doi:10.1016/j.imu.2019.100250
32. Nakajima N, Yoshizawa A, Nakajima T, et al. GATA6‐positive lung adenocarcinomas are associated with invasive mucinous adenocarcinoma morphology, hepatocyte nuclear factor 4α expression, and KRAS mutations. Histopathology. 2018. doi:10.1111/his.13500
33. Chia NY, Deng N, Das K, et al. Regulatory crosstalk between lineage-survival oncogenes KLF5, GATA4 and GATA6 cooperatively promotes gastric cancer development. Gut. 2015;64(5):707–719. doi:10.1136/gutjnl-2013-306596
34. Song Y, Tian T, Fu X, et al. GATA6 is overexpressed in breast cancer and promotes breast cancer cell epithelial mesenchymal transition by upregulating slug expression. Exp Mol Pathol. 2015;99(3):617–627. doi:10.1016/j.yexmp.2015.10.005
35. Fantini S, Salsi V, Reggiani L, et al. The miR-196b miRNA inhibits the GATA6 intestinal transcription factor and is upregulated in colon cancer patients. Oncotarget. 2017;8(3):4747–4759. doi:10.18632/oncotarget.13580
36. Tian F, Chen J, Zheng S, et al. miR-124 targets GATA6 to suppress cholangiocarcinoma cell invasion and metastasis. BMC Cancer. 2017;17(1):175. doi:10.1186/s12885-017-3166-z
37. Novellasdemunt L, Antas P. Li V S W. Targeting Wnt signaling in colorectal cancer. Ajp Cell Physiol. 2015;
38. van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–260. doi:10.1146/annurev.physiol.010908.163145
39. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi:10.1038/nrc822
40. De Lau W, Peng WC, Gros P, et al. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28(4):305–316. doi:10.1101/gad.235473.113
41. Qi L, Sun B, Liu Z, et al. Dickkopf-1 inhibits epithelial-mesenchymal transition of colon cancer cells and contributes to colon cancer suppression. Cancer Sci. 2012;103(4):828–835. doi:10.1111/j.1349-7006.2012.02222.x
42. Yang B, Mao L, Li Y, et al. β-catenin, leucine-rich repeat-containing G protein-coupled receptor 5 and GATA-binding factor 6 are associated with the normal mucosa-adenoma-adenocarcinoma sequence of colorectal tumorigenesis. Oncol Lett. 2017. doi:10.3892/ol.2017.7566
43. Rønnov-Jessen L, Bissell MJ. Breast cancer by proxy: can the microenvironment be both the cause and consequence? Trends Mol Med. 2009;15(1):5–13. doi:10.1016/j.molmed.2008.11.001
44. Monteith GR, Davis FM, Roberts-Thomson SJ. Calcium channels and pumps in cancer: changes and consequences. J Biol Chem. 2012;287(38):31666–31673. doi:10.1074/jbc.R112.343061
45. Qiu Y, Michalak M. Transcriptional control of the calreticulin gene in health and disease. Int J Biochem Cell Biol. 2009;41(3):
46. Ihara Y, Inai Y, Ikezaki M, et al. Alteration of integrin-dependent adhesion and signaling in EMT-like MDCK cells established through overexpression of calreticulin. J Cell Biochem. 2011;112:2518–2528. doi:10.1002/jcb.23176
47. Kwon MS, Park CS, Choi K, et al. Calreticulin couples calcium release and calcium influx in integrin-mediated calcium signaling. Mol Biol Cell. 2000;11(4):1433–1443. doi:10.1091/mbc.11.4.1433
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.