Back to Journals » OncoTargets and Therapy » Volume 12
miR-942 promotes proliferation and metastasis of hepatocellular carcinoma cells by inhibiting RRM2B
Authors Zhang Q, Zhu B, Qian J, Wang K, Zhou J
Received 4 March 2019
Accepted for publication 16 July 2019
Published 11 October 2019 Volume 2019:12 Pages 8367—8378
DOI https://doi.org/10.2147/OTT.S207549
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Qifan Zhang,1,* Bili Zhu,2,* Jianping Qian,1 Kai Wang,1 Jie Zhou1
1Department of Hepatobiliary Surgery, Nanfang Hospital, Southern Medical University, Guangzhou 510515, People’s Republic of China; 2Huiqiao Medical Center, Nanfang Hospital, Southern Medical University, Guangzhou 510515, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie Zhou
Huiqiao Medical Center, Nanfang Hospital, Southern Medical University, Guangzhou Avenue No.1838, Guangzhou 510515, Guangdong Province, People’s Republic of China
Tel +86 0 206 278 7180
Email [email protected]
Background: Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death. MicroRNA-942 (miR-942) plays a critical role in promoting proliferation and metastasis of cancer cells and is associated with poor prognosis in some types of cancers. However, the prognostic value of miR-942 and its functional role in HCC remain unclear.
Materials and methods: Real-time PCR (RT-PCR) was used to detect the expression of miR-942 in HCC tissues and adjacent normal liver tissues. Next, the correlations between miR-942 expression and clinicopathological parameters including the survival rate were analyzed. Interaction between miR-942 and ribonucleotide reductase regulatory TP53 inducible subunit M2B (RRM2B) was determined by RT-PCR, Western blot and luciferase assay. The biological influence of miR-942 on HCC cell lines was studied using CCK-8 assay, colony formation assay and transwell assay in vitro. Western blot and RT-PCR were used to analyze the change of downstream genes after miR-942 mimics transfection.
Results: miR-942 was significantly up-regulated in HCC. Its high expression was associated with serum alanine transaminase level (P=0.0350), tumor size (P=0.0195), T stage (P=0.0045) and lymphatic metastasis (P=0.0013). High expression of miR-942 was associated with shorter overall survival and disease-free survival time of HCC patients. RRM2B was validated as a target gene of miR-942. miR-942 mimics markedly promoted the malignant phenotypes of Huh7 and MHCC97H cell lines, while its inhibitor had the opposite effect. miR-942 can regulate the downstream genes of RRM2B including Egr-1 and PTEN, markers of epithelial-mesenchymal transition and matrix metalloproteinases.
Conclusion: miR-942 may serve as a potential biomarker for HCC and its inhibitor may be a therapeutic agent for the treatment of this deadly disease.
Keywords: miR-942, RRM2B, hepatocellular carcinoma, cancer
Introduction
Liver cancer is one of the most common malignancies. According to statistics, in every year, there were about 800,000 new cases of liver cancer and the number of deaths due to liver cancer was approximately 700,000 all over the world, and China accounted for 50% of the total number of new cases and deaths.1,2 Hepatocellular carcinoma (HCC) is the main type of liver cancer, which accounts for 70–90% of liver cancer.1,2 However, the underlying pathological mechanisms of HCC progression remain unclear and there is a lack of effective treatment for it. At present, surgical resection is still the preferred method for the treatment of HCC. However, the high 5-year recurrence and metastasis rate of patients after radical resection are the difficult problems in the clinical treatment of HCC. Therefore, studying the mechanism of HCC at the molecular level and looking for effective treatment measures have become the focus and difficulty in the research of HCC.
microRNA (miR) is a non-encoding single-stranded RNA consisting of about 20 nucleotides.3 Accumulating evidence suggests a role for microRNAs in human carcinogenesis as novel tumor suppressors or oncogenes.4–6 miR-942 is a newly discovered miR, and previous studies have reported that it can regulate the development of a variety of tumors.7–9 However, there are few reports on the expression, clinical significance and function of miR-942 in HCC.
Ribonucleotide Reductase (RR), ubiquitous in a variety of cells, participates in the process of ribose nucleotide reduction into deoxyribose and plays a central role in nucleoside metabolism.10 It is the only enzyme that transforms ribose nucleotides into deoxyribose nucleotides.10 Deoxyribonucleotides are raw materials for DNA synthesis and repair, so this enzyme is a rate-limiting enzyme for repair and synthesis in DNA pathway.11 The whole structure of RR includes large-subunit α and small-subunit β, and only the formation of α2β2 heterotetramer structure can be activated.11,12 In human cells, RR consists of 3 subunits, 1 large-subunit [ribonucleotide reductase M1 (RRM1)] and 2 small-subunits (RRM2 and RRM2B).10 Because RR plays a key role in the synthesis of DNA, it has important effects on the biological behaviors of cancer cells. Existing studies have indicated RRM1, RRM2 and RRM2B, as oncogenes or tumor suppressor, have been implicated in multiple malignancies.13–16 Specifically, RRM2B can inhibit the metastasis of HCC by activating Egr-1/PTEN/Akt1 signaling pathway, suggesting that RRM2B plays a role of tumor suppressor gene in HCC.17
In this proof-of-principle study to identify potential biomarkers for HCC, we carried out the study on the expression of miR-942 and its clinical significance for HCC patients. We found that miR-942 was highly expressed in HCC, whose association with poor prognosis of patients was verified in the present study. RRM2B was predicted as a target gene of miR-942. Taken together, miR-942 could repress the expressions of Egr-1 and PTEN via regulating RRM2B. Our results provide robust evidences that miR-942 functions as a potent enhancer of HCC cell proliferation via modulating RRM2B.
Methods and materials
Patient specimens and cell lines
The study was approved by the Ethics Committee of the Nanfang Hospital of Southern Medical University. Sixty samples of HCC tissues and their corresponding adjacent tissues were obtained from the Southern Medical University. All patients provided written informed consent for their tissues to be used in this study, which was in accordance with the Declaration of Helsinki. All of the specimens were confirmed by histopathological examination by two pathologists. 293T and HCC cell lines, including Hep3B, Huh7, SMMC-7721, Bel-7402, MHCC97L and MHCC97H were obtained from our lab conservation. Immortalized normal liver cell line L02 was purchased from the Chinese Academy of Sciences Committee Type Culture Collection cell bank. The use of the cell lines was also approved by the Ethics Committee of the Nanfang Hospital of Southern Medical University. Cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen) in humidified air at 37°C with 5% CO2. The media were supplemented with 10% fetal bovine serum (10% FBS), 100 U/mL penicillin and 100 mg/mL streptomycin (Invitrogen, Shanghai, China). In subsequent experiments, cells in logarithmic growth phase were used. miR-942 mimics, inhibitors and irrelevant sequences (negative control group) were transfected into cells using liposome Lipofectamine 3000, which referred to the reagent instructions for the procedure.
Real-time quantitative PCR
The total RNA of tissues and cells was extracted by Trizol method. For the quantification of miR-942, reverse transcription reaction was carried out using TapMan MicroRNA Reverse Transcription kit (Invitrogen, Shanghai, China). The reaction conditions were: 16°C for 30 mins, 42°C for 30 mins and 85°C for 5 mins. The quantitative amplification reaction was carried out using the TapMan MicroRNA Assays Real-time PCR kit (Invitrogen, Shanghai, China). The reaction conditions were: 95°C for 5 mins; 95°C for 15 s, 62°C for 30 s, for a total of 40 cycles. For the quantification of mRNA, reverse transcription reaction was using High Capacity cDNA Reverse Transcription Kit (Thermo Fisher, Shanghai, China). RT-PCR analyses were performed with the SYBR Green PCR Master Mix (TaKaRa, Otsu, Shiga, Japan) in accordance with the manufacturer’s instructions. The primers were listed in the supplementary materials. Three replicate experiments were conducted for each test index, and the expression levels of miR-942, RRM2B and other genes were relatively quantified by the 2−ΔΔCt method.
Reporter gene assay
293T cells were seeded in 24-well plates (1.5×105/well), and the cells were grown to about 80%. miR-NC + pGL3-RRM2B-wt, miR-942 mimics + pGL3-RRM2B-wt, miR-942 mimics + pGL3-RRM2B-mut were co-transfected into 293T cells, respectively, and fresh medium was replaced after 6 hrs. After 48 hrs of transfection, each group of cells was lysed and manipulated according to the Dual-Luciferase Reporter Assay System instructions, and the fluorescence intensity of each group of cells was expressed as the ratio of firefly luciferase activity to Renilla luciferase activity.
Proliferation experiment
Each group of cells was trypsinized with 0.25% trypsin. After resuspending the cell suspension, the cells were seeded in a 96-well plate at a density of 1000/well, and the volume was 100 μL per well, and the culture was continued in an incubator. Cell viability was measured by CCK-8 method on days 1, 2, 3, 4 and 5, respectively. The cell growth curve was plotted with time as the horizontal axis and 490 nm absorbance as the vertical axis.
Colony formation experiment
Each group of cells was trypsinized with 0.25% trypsin. The cell suspension was prepared in complete medium, seeded in a 12-well plate at 1000/well, and each group of cells was made into 3 replicate wells, and then placed in an incubator to continue the culture. After the culture for 14 days, the culture was terminated, and the culture solution was aspirated, washed three times with PBS solution, and then stained with 0.1% crystal violet and 10% methanol at room temperature for 20 mins, and then washed with running water slowly. After drying, the number of colonies visible to the naked eye was counted.
Transwell experiments
The transwell chamber was using a polycarbonate filter of 8 μm size (Corning, Beijing, China) for migration experiments. Each group of cells was made into a cell suspension of 1*105/mL density in serum-free medium, and 200 μL of the cell suspension was inoculated into the upper chamber of the Transwell chamber, and 500 μL of 10% fetal bovine serum was added to the lower chamber. There are 3 duplicate holes in each group. After incubating in an incubator for 24 hrs, the upper uncoated cells were gently wiped off with a cotton swab. The cells were fixed with 4% paraformaldehyde for 10 mins and stained with 0.1% crystal violet for 10 mins. The number of cells was then counted under a microscope. The chamber of the invasion experiment was covered with matrigel, and the other steps were the same as the migration experiment.
Western blot
Anti-RRM2B antibody (abcam, ab8105, 1:1000), anti-Vimentin antibody (abcam, ab92547, 1:500), anti-N-cadherin antibody (abcam, ab18203, 1:1000), anti-ZO-1 antibody (abcam, ab96587, 1:1000), anti-E-cadherin (abcam, ab40772, 1:500), anti-MMP-2 antibody (abcam, ab37150, 1:500), anti-MMP-9 antibody (abcam, ab137867, 1:500), anti-PTEN antibody (abcam, ab170941, 1:1000), anti-Egr-1 antibody (abcam, ab55160, 1:1000) and anti-GAPDH antibody (abcam, ab181602, 1:100) were used in Western blot. The cells were lysed on ice for 30 mins using RIPA lysate, and the protein concentration was quantified by the BCA protein quantification kit (Thermo Fisher, Shanghai, China). Gel electrophoresis was performed using 12% SDS-PAGE gel, and the protein loading per lane was 20 μg. The gel was electrophoresed at 60 V for 1 hr, and the gel was separated by electrophoresis at 100 V for 2 hrs, and transferred to nitrocellulose membranes by a conventional wet transfer method for 90 mins. Then the membranes were blocked with 5% defatted milk in TBST for 1 hr, and added to the corresponding primary antibodies at 4°C overnight. After washed for 3 times, the membranes were incubated with corresponding secondary antibodies at room temperature for 2 hrs. At last, the membranes were washed 3 times with TBST, and the signals were detected by chemiluminescence phototope-HRP kit (Pierce Biotechnology, USA) according to the manufacturer’s instructions.
Statistical analysis
Kmplotter (http://kmplot.com) was used to conduct Kaplan-Meier survival analysis.18 The correlation between miR-942 expression and the clinicopathologic features was analyzed by Chi-square test. All experiments were repeated 3 times and the experimental data were expressed as mean ± standard deviation (x ± s). Statistical analysis was performed using SPSS 17.0 software, and P<0.05 was considered statistically significant.
Results
miR-942 is highly up-regulated in HCC tissues and associated with worse prognosis
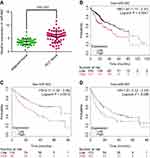
The expression of miR-942 was investigated in 60 pairs of HCC tissue and adjacent normal tissues by qRT-PCR. The results revealed that miR-942 was significantly (P<0.001) up-regulated in the HCC tissues relative to the normal tissues (Figure 1A). To investigate the clinical significance of up-regulation of miR-942 in HCC, we equally divided the 60 HCC samples into two groups (high expression vs low expression) according to the results of qRT-PCR, and carried out χ2 analysis to determine the relationship between miR-942 expression and clinicopathologic features. As shown in Table 1, statistical analysis indicated that there was a significantly positive correlation of miR-942 expression with higher serum alanine transaminase (ALT) level (P=0.0350), larger tumor size (P=0.0195), higher T stage (P=0.0045) and higher probability of lymphatic metastasis (P=0.0013). These results implied that miR-942 may promote tumor growth and metastasis of HCC. Importantly, Kaplan-Meier survival analysis showed the patient survival time (both overall survival time and disease-free survival time) in miR-942 high expression group was significantly shorter than in the low expression group, and this conclusion was validated in 2 different cohorts (Figure 1B–D). These results further implied that miR-942 positively relates to tumor progression. We also detected miR-942 expression in HCC cell lines (Hep3B, Huh7, SMMC-7721, Bel-7402, MHCC97 and MHCC97H) and an immortal normal liver cell line (L02) by qRT-PCR. As illustrated (Figure 2), the expression levels of miR-942 were significantly higher in most HCC cell lines, especially MHCC97L and MHCC97H, which were considered as cancer cell lines with high potential of metastasis. Collectively, these data suggested that miR-942 is highly up-regulated in HCC and associated with worse prognosis.
 |
Table 1 Correlation between the miR-942 expression and the clinicopathologic features of hepatocellular carcinoma |
miR-942 modulates the proliferation of HCC cells
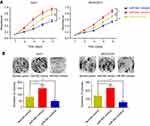
To decipher the role of miR-942, we, respectively, transfected its mimics and inhibitor into Huh7 and MHCC97H cells, and then observed the change of phenotypes of the cells. CCK-8 assays demonstrated that miR-942 caused significant increase in the viability of both Huh7 and MHCC97H cells, while miR-942 inhibitor had the opposite effects (Figure 3A). We also conducted colony formation assays. As shown (Figure 3B), suppression of miR-942 caused significant decline in the colony formation of HCC cells, while its mimics promoted the colony formation ability of HCC cells. Taken together, we concluded that the up-regulation of miR-942 in HCC tissues could enhance the growth of tumor.
miR-942 modulates migration and invasion of HCC cells by regulating epithelial-mesenchymal transition and matrix metalloproteinases
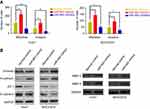
In order to further assess the effects of miR-942 dysregulation on HCC cells, we conducted transwell assays to determine the ability of migration and invasion, which were two key factors of metastasis. As shown, overexpression of miR-942 led to significantly enhanced migration, invasion and the ability of wound healing in both Huh7 and MHCC97H cells; conversely, the suppression of miR-942 inhibited the migration, invasion and wound healing (Figure 4A, Figure S1A and B). Epithelial-mesenchymal transition (EMT) endows cells with migratory and invasive properties and is thought to play crucial roles in tumor progression and metastasis; matrix metalloproteinases (MMPs) are involved in the degradation of extracellular matrix, and are related to the development, invasion, metastasis and occurrence of cancers.19,20 So we also detected the markers of the EMT and MMP-2/9 by Western blot after miR-942 overexpression. As shown (Figure 4B), miR-942 mimics induced the expression of mesenchymal markers N-cadherin and vimentin, MMP-2 and MMP-9, while the epithelial markers E-cadherin and ZO-1 were inhibited. Collectively, these results suggested that miR-942 was a modulator of HCC metastasis.
miR-942 directly targets 3ʹUTR of RRM2B and negatively regulated its expression
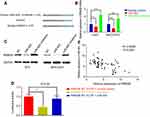
In this study, we searched TargetScan database to look for potential novel target genes of miR-942, and RRM2B caught our attention (Figure 5A), for that the decrease of RRM2B in HCC is reported to contribute to poor prognosis of patients.17 We demonstrated that miR-942 transfection into HCC cells resulted in a profound reduction of RRM2B expression at both mRNA and protein levels; however, miR-942 inhibitors had an opposite effect. Consistent results were obtained from Huh7 and MHCC97H cells (Figure 5B and C), which indicated the negative regulation of RRM2B by miR-942. RRM2B was further confirmed to be the target of miR-942 by luciferase reporter assay (Figure 5D). Additionally, expressions of RRM2B and miR-942 were inversely correlated in the 60 HCC samples (Figure 3E). Taken together, our data suggested that miR-942 may exert oncogenic function via inhibiting RRM2B in HCC.
Up-regulation of mir-942 may be involved in cancer progression via regulating RRM2B/Egr-1/PTEN pathway
It is reported that RRM2B inhibits HCC progression through regulation of the early growth response protein 1 (Egr-1)/phosphatase and tensin homolog (PTEN)/Akt pathway.17 As mentioned previously, we had proven that RRM2B was a target gene of miR-942. So we made a hypothesis that miR-942 could indirectly regulate the expression of Egr-1 and PTEN, and in turn change the expression of EMT makers and MMPs. Consistent with our expectations, in both Huh7 and MHCC97H cells, transfection of miR-942 mimics reduced the expression levels of Egr-1 and PTEN on protein levels, while transfection of miR-942 inhibitors increased the expression levels of Egr-1 and PTEN (Figure 6A). What is more, in Huh7 cell, qRT-PCR showed that miR-942 mimics inhibited the expression of PTEN, Egr-1, E-cadherin, ZO-1, TIMP-1 and TIMP-3, while inducing the expression of vimentin, N-cadherin, MMP-2 and MMP-9, and the effects of miR-942 mimics could be revered by co-transfection of RRM2B (Figure 6B). In contrast, compared with miR-942 mimics, miR-942 inhibitor had opposite effects (Figure 6B). Based on these results, we considered that miR-942 promoted HCC progression through regulating RRM2B/Egr-1/PTEN pathway.
Discussion
The oncogenic role of miR-942 has been reported in several studies. The expression of miR-942 in bladder cancer tissues is significantly higher than that of normal tissues, and its expression level is closely related to clinical staging.7 In metastatic renal carcinoma, miR-942 can increase the secretion of MMP-9 and VEGF, promoting the angiogenesis of tumor tissue and the migration of cancer cells, enhancing the resistance of cancer cells to schunitinib.8 In colorectal cancer, negatively regulated by LINC00675, miR-942 is involved in regulating proliferation and metastasis of cancers.9 In esophageal squamous cell carcinoma, miR-942 can up-regulate the activity of wnt/β-catenin signaling pathway by targeting genes such as sFRP4, GSK3β and TLE1, to promote cancer stem cell-like traits.21 miR-942 can also inhibit the endogenous expression level of ISG12a, thereby inhibiting TRAIL-induced apoptosis of cancer cells.22 Consistent with previous studies, our results validated the involvement of miR-942 in HCC. It is worth mentioning that we further demonstrated the detailed mechanism of HCC, that is, miR-942 could inhibit the expression of tumor suppressor genes RRM2B and PTEN to facilitate HCC progression.
RRM1, RRM2 and RRM2B have been well documented to be involved in diverse cancers. For instance, RRM1 might inhibit the migration, invasion and metastasis of lung cancer cells through the expression of PTEN.14 It is also found that in patients with non-small cell lung cancer, RRM1 expression is closely related to the expression of PTEN and RRM2, and patients with high expression of RRM1 have better prognosis.14,23 Therefore, RRM1 is considered to be a suppressor gene for tumor metastasis. Several studies have shown that RRM2 may play a role similar to oncogenes in the development and progression of tumors. RRM2 expression is reported to elevate in the bulk of cancers, like gastric cancer, colon cancer and esophageal cancer, which is verified to be closely linked to tumor staging and patient prognosis.15,24,25 RRM2B was originally used as the target gene for tumor suppressor protein p53, also known as p53R2. The expression of RRM2B is found to be positively correlated with unfavorable prognosis in studies on esophageal cancer, oral cancer, small cell lung cancer and others.26–28
However, in colon cancer and HCC, it was found that the expression of RRM2B is down-regulated, and was negatively correlated with metastasis, and in these tumors, RRM2B is believed to be a tumor suppressor.17,29 Our research confirms that the abnormally high expression of miR-942 is one of the mechanisms for the decreased expression of RRM2B in HCC. Combined with previous reports, We assume that miR-942/RRM2B/Egr-1/PTEN pathway may be an important mechanism in the progression of HCC. Of course, the abnormal expression of miR-942 and RRM2B may affect malignant phenotypes of tumor cells in other ways. It has been reported that RRM2B inhibits the phosphorylation level of extracellular signal-regulated kinase by interacting with mitogen-activated protein kinase 2, up-regulates p21 expression and down-regulates cyclin D1 expression, thus inhibiting tumor cell proliferation.30,31 In HCC, whether there is a similar mechanism needs to be further explored in the future.
In conclusion, our data show that miR-942 expression is higher in HCC and associated with worse prognosis. By inhibiting RRM2B/Egr-1/PTEN, miR-942 accelerates HCC proliferation and metastasis. Thus, miR-942 may serve as a potential biomarker for HCC and its inhibitor may be a therapeutic agent for the treatment of this deadly disease. In the future, in vivo studies are needed to further verify its role in HCC and other target genes should be screened and validated.
Ethics statement
Our study was approved by the ethics review board of the Nanfang Hospital.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgment
This study is supported by the Natural Science Foundation of Guangdong Province (No. 2017A030313684) and Guangzhou Science and Technology Project (No. 201400000001-3 and 201508020262).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
3. Yuan Y, Weidhaas JB. Functional microRNA binding site variants. Mol Oncol. 2018. doi:10.1002/1878-0261.12421
4. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer – a brief overview. Adv Biol Regul. 2015;57:1–9. doi:10.1016/j.jbior.2014.09.013
5. Yang N, Zhu S, Lv X, Qiao Y, Liu YJ, Chen J. MicroRNAs: pleiotropic regulators in the tumor microenvironment. Front Immunol. 2018;9:2491. doi:10.3389/fimmu.2018.02491
6. Xu X, Tao Y, Shan L, et al. The role of MicroRNAs in hepatocellular carcinoma. J Cancer. 2018;9(19):3557–3569. doi:10.7150/jca.26350
7. Zhang ZC, Huang Y, Wang XJ, Wang M, Ma LL. Expression of circulating microRNAs in patients with bladder urothelial carcinoma. Beijing Da Xue Xue Bao Yi Xue Ban. 2013;45(4):532–536.
8. Prior C, Perez-Gracia JL, Garcia-Donas J, et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS One. 2014;9(1):e86263. doi:10.1371/journal.pone.0086263
9. Shan Z, An N, Qin J, Yang J, Sun H, Yang W. Long non-coding RNA Linc00675 suppresses cell proliferation and metastasis in colorectal cancer via acting on miR-942 and Wnt/β-catenin signaling. Biomed Pharmacother. 2018;101:769–776. doi:10.1016/j.biopha.2018.02.123
10. Aye Y, Li M, Long MJ, Weiss RS. Ribonucleotide reductase and cancer: biological mechanisms and targeted therapies. Oncogene. 2015;34(16):2011–2021. doi:10.1038/onc.2014.155
11. Shao J, Liu X, Zhu L, Yen Y. Targeting ribonucleotide reductase for cancer therapy. Expert Opin Ther Targets. 2013;17(12):1423–1437. doi:10.1517/14728222.2013.840293
12. Wang X, Zhenchuk A, Wiman KG, Albertioni F. Regulation of p53R2 and its role as potential target for cancer therapy. Cancer Lett. 2009;276(1):1–7. doi:10.1016/j.canlet.2008.07.019
13. Jordheim LP, Sève P, Trédan O, Dumontet C. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 2011;12(7):693–702. doi:10.1016/S1470-2045(10)70244-8
14. Cerqueira NM, Fernandes PA, Ramos MJ. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat Anticancer Drug Discov. 2007;2(1):11–29.
15. Zheng S, Wang X, Weng YH, et al. siRNA knockdown of RRM2 effectively suppressed pancreatic tumor growth alone or synergistically with doxorubicin. Mol Ther Nucleic Acids. 2018;12:805–816. doi:10.1016/j.omtn.2018.08.003
16. Chen J, Li S, Xiao Y, et al. p53R2 as a novel prognostic biomarker in nasopharyngeal carcinoma. BMC Cancer. 2017;17(1):846. doi:10.1186/s12885-017-3858-4
17. Tian H, Ge C, Li H, et al. Ribonucleotide reductase M2B inhibits cell migration and spreading by early growth response protein 1-mediated phosphatase and tensin homolog/Akt1 pathway in hepatocellular carcinoma. Hepatology. 2014;59(4):1459–1470. doi:10.1002/hep.26929
18. Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi:10.1038/s41598-018-27521-y
19. Tania M, Khan MA, Fu J. Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumour Biol. 2014;35(8):7335–7342. doi:10.1007/s13277-014-2163-y
20. Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhibitors in cancer therapy: turning past failures into future successes. Mol Cancer Ther. 2018;17(6):1147–1155. doi:10.1158/1535-7163.MCT-17-0646
21. Ge C, Wu S, Wang W, et al. miR-942 promotes cancer stem cell-like traits in esophageal squamous cell carcinoma through activation of Wnt/β-catenin signalling pathway. Oncotarget. 2015;6(13):10964–10977. doi:10.18632/oncotarget.3696
22. Liu N, Zuo C, Wang X, et al. miR-942 decreases TRAIL-induced apoptosis through ISG12a downregulation and is regulated by AKT. Oncotarget. 2014;5(13):4959–4971. doi:10.18632/oncotarget.2067
23. Bepler G, Sharma S, Cantor A, et al. RRM1 and PTEN as prognostic parameters for overall and disease-free survival in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22(10):1878–1885. doi:10.1200/JCO.2004.12.002
24. Zhong Z, Cao Y, Yang S, Zhang S. Overexpression of RRM2 in gastric cancer cell promotes their invasiveness via AKT/NF-κB signaling pathway. Pharmazie. 2016;71(5):280–284.
25. Fang Z, Gong C, Liu H, et al. E2F1 promote the aggressiveness of human colorectal cancer by activating the ribonucleotide reductase small subunit M2. Biochem Biophys Res Commun. 2015;464(2):407–415. doi:10.1016/j.bbrc.2015.06.103
26. Okumura H, Natsugoe S, Yokomakura N, et al. Expression of p53R2 is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12(12):3740–3745. doi:10.1158/1078-0432.CCR-05-2416
27. Yanamoto S, Kawasaki G, Yoshitomi I, Mizuno A. Expression of p53R2, newly p53 target in oral normal epithelium, epithelial dysplasia and squamous cell carcinoma. Cancer Lett. 2003;190(2):233–243. doi:10.1016/s0304-3835(02)00588-8
28. Uramoto H, Sugio K, Oyama T, Hanagiri T, Yasumoto K. P53R2, p53 inducible ribonucleotide reductase gene, correlated with tumor progression of non-small cell lung cancer. Anticancer Res. 2006;26(2A):983–988.
29. Liu X, Zhou B, Xue L, et al. Metastasis-suppressing potential of ribonucleotide reductase small subunit p53R2 in human cancer cells. Clin Cancer Res. 2006;12(21):6337–6344. doi:10.1158/1078-0432.CCR-06-0799
30. Piao C, Jin M, Kim HB, et al. Ribonucleotide reductase small subunit p53R2 suppresses MEK-ERK activity by binding to ERK kinase 2. Oncogene. 2009;28(21):2173–2184. doi:10.1038/onc.2009.84
31. Zhang K, Wu J, Wu X, et al. p53R2 inhibits the proliferation of human cancer cells in association with cell-cycle arrest. Mol Cancer Ther. 2011;10(2):269–278. doi:10.1158/1535-7163.MCT-10-0728
Supplementary material
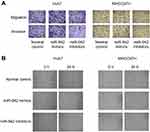 |
Figure S1 (A) Transwell assay was used to detect migration and invasion; (B) Would healing assay was used to detect migration. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.