Back to Journals » OncoTargets and Therapy » Volume 13
miR-873-5p Inhibits Cell Migration and Invasion of Papillary Thyroid Cancer via Regulation of CXCL16
Authors Wang Z, Liu W, Wang C, Ai Z
Received 23 April 2019
Accepted for publication 27 December 2019
Published 4 February 2020 Volume 2020:13 Pages 1037—1046
DOI https://doi.org/10.2147/OTT.S213168
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Zhenglin Wang,* Wei Liu,* Cong Wang, Zhilong Ai
Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhilong Ai
Department of General Surgery, Zhongshan Hospital, Fudan University, 180 Fenglin Road, Xuhui District, Shanghai 200032, People’s Republic of China
Email [email protected]
Aim: Papillary thyroid cancer (PTC) is the most common type of thyroid cancer with an increasing morbidity. MicroRNAs (miRNAs) play the pivotal roles in PTC occurrence and development. The aim of this study was to investigate the biological functions of miR-873-5p and its underlying molecular mechanisms in PTC.
Methods: Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis was performed to detect miR-873-5p expressions in PTC tissues and cell lines. The target gene of miR-873-5p was predicted by TargetScan and confirmed by dual-luciferase reporter assay. Furthermore, cell proliferation, migration and invasion were assessed by CCK-8, wound healing assay and transwell assay, respectively. Additionally, the expressions of CXCL16, MMP1, MMP9 and MMP13 were measured by RT-qPCR and Western blot methods, and p65, Rel-B and their phosphorylation levels were examined by Western blot.
Results: We found that miR-873-5p expression was downregulated in PTC tissues and cell lines. Moreover, CXCL16 was identified as a target of miR-873-5p, and its expression was upregulated in PTC tissues and cells at both mRNA and protein levels. Functionally, overexpression of miR-873-5p inhibited PTC cell proliferation, migration and invasion, while co-transfection of CXCL16 overexpression plasmid reversed the anti-tumor behaviors induced by miR-873-5p. In addition, miR-873-5p overexpression suppressed the phosphorylation of p65 and Rel-B, and decreased the mRNA and protein expression of MMP1, MMP9 and MMP13, while overexpression of CXCL16 partially abrogated the effects of miR-873-5p.
Conclusion: MiR-873-5p functions as a tumor suppressor in PTC by inhibiting the proliferation, migration and invasion of the PTC cells via targeting CXCL16. These findings might provide a potential novel target for the therapy of PTC.
Keywords: papillary thyroid cancer, miR-873-5p, CXCL16, migration, invasion, proliferation
Introduction
Thyroid cancer is a common endocrine malignancy with an increasing morbidity and young trend in recent years.1 Thyroid cancer can be divided into four types, including papillary, follicular, medullary and anaplastic tumor. Papillary thyroid cancer (PTC) is the major histological subtype of thyroid cancer, which accounts for 80–90% of all thyroid carcinoma.2 While most of the patients with PTC have an excellent therapeutic response and long-term survival,3 some patients, especially in older patients, are associated with lymph node metastasis, which results in an increased risk of locoregional recurrence and poor prognosis.4 Moreover, because of no obvious clinical symptoms, PTC is difficult to diagnose at early stage,5 and surgical management remains controversial.6 Therefore, it is necessary to find novel biomarkers and therapeutic targets for PTC early diagnosis and treatment.
MicroRNAs (miRNAs) are a class of endogenous, approximately 22nt in length and single-strand RNAs, which play important roles in regulating the target mRNAs via cleavage or translational repression.7 They negatively regulate gene expression by pairing the 3ʹUTR site of target mRNA. MiRNAs have been reported to be involved in cellular processes, such as inflammation, cell differentiation, cell-cycle regulation, apoptosis, migration and metastasis.8 In various human cancers, miRNAs control the expression of mRNAs to regulate tumor growth, invasion, angiogenesis and immune evasion.9 It is reported that miRNAs were involved in every stage of PTC, including tumor diagnosis, prognosis, treatment and surveillance.10 According to the small RNA deep-sequencing data, miR-873-5p expression in PTC is downregulated in tumors compared to normal samples.11 However, the roles of miR-873-5p in PTC and the underlying molecular mechanisms remain unknown.
In the present study, we confirmed the down-regulation of miR-873-5p in PTC. We also explored the regulatory roles of miR-873-5p in PTC cells in vitro. Furthermore, the target gene and the underlying molecular mechanisms of miR-873-5p in PTC were investigated. This study would provide a new target for PTC treatment.
Materials and Methods
Clinical Tissue Samples
A total of 30 pairs of PTC tissues and the adjacent normal tissues (at least 2cm from the PTC tumor tissues) were obtained from Zhongshan Hospital. The mean age of the patients was 45.83±12.71, and male/female was 9/21. All tissue samples were maintained at −80°C until further use. The Ethics Committee of Zhongshan Hospital approved this study protocol (Approval NO. 20170523), and written informed consent was obtained from all participants in accordance with the Declaration of Helsinki.
Cell Culture
Human PTC cell lines KTC-1, TPC-1, BCPAP, K1, BHP10-3 and normal thyroid epithelial cell line Nthy-ori3-1, which were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China), were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin and 100 μg/mL of streptomycin (All purchased from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were incubated in an incubator at 37°C with 5% CO2.
Prediction of Target Genes and Dual-Luciferase Reporter Assay
Targetscan (http://www.targetscan.org/vert_72/) was used to identify the target genes of miR-873-5p. The wild-type 3ʹUTR fragment of CXCL16 cDNA and mutant CXCL16 3ʹUTR were amplified and inserted into pmirGLO vector (Promega, Madison, WI, USA). HEK 293T cells were incubated in 24-well plates and co-transfected with pmirGLO-CXCL16-WT or pmirGLO-CXCL16-mutant together with miR-873-5p mimics or miR-NC mimics, which was performed by using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.). Forty-eight hours later, the relative luciferase activity was measured by Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The results were quantified by normalizing the firefly activity to Renilla activity.
Transfection
For the investigation of miR-873-5p on the behaviors of PTC cells, miR-873-5p mimics and corresponding negative control (miR-NC mimics) were synthesized by GenePharma (Shanghai, China). For the overexpression of CXCL16, CXCL16 cDNA was cloned into pcDNA3.1 vector (Thermo Fisher Scientific, Inc.). TPC-1 and K1 cells were seeded into 6-well plates 1 day before transfection and were divided into four groups, including control, miR-NC mimics, miR-873-5p mimics and miR-873-5p mimics + pcDNA3.1-CXCL16 groups. The transfection was carried out by using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, Inc.). At 6 h post-transfection, culture media were replaced with complete medium and cells were continued to incubate for another 48 h.
RNA Isolation and RT-qPCR
Total RNA from tissues and cells was extracted by using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.). To detect miR-873-5p expression, reverse transcription and qPCR were performed by using TransScript Green miRNA Two-step qRT-PCR SuperMix (TransGen, Beijing, China). For mRNA expression, RNA was reverse transcribed into cDNA using TransScript First-Strand cDNA Synthesis superMix (TransGen) and qPCR was conducted by TransStart Green qPCR SuperMix (TransGen). The qPCR reaction was composed of 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and 60°C for 30 s on ABI Prism 7700 (Applied Biosystems, Foster City, CA, USA). The miR-873-5p expression was normalized to U6 and other mRNA expression was normalized to GAPDH. The fold change was calculated using the 2–ΔΔCt method.12 The sequences of the primers were miR-873-5p, forward 5ʹ-GCAGGAACUUGUGAGUCUCCU-3ʹ and reverse 5ʹ-AGGAGACUCACAAGUUCCUGC-3ʹ; U6, forward 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and reverse 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ; CXCL16, forward 5ʹ-ACTCAGCCAGGCAATGGCAAC-3ʹ and reverse 5ʹ-GGTATTAGAGTCAGGTGCCAC-3ʹ; GAPDH, forward 5ʹ-CTGGTCACCAGGGCTGCTTTT-3ʹ and reverse 5ʹ-CATGAGGTCCACCACCCTGTT-3ʹ.
Western Blot
Total protein was extracted by ProteoPrep Total Extraction Sample Kit (Sigma-Aldrich, St Louis, MO, USA) and the concentration was quantified using BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.). Then, the protein was separated by 10% SDS-PAGE and transferred onto PVDF membranes (Bio-Rad, Hercules, CA, USA). The membranes were blocked with TBST containing 5% non-fat milk, followed by incubated with primary antibodies overnight at 4°C and secondary antibody for 1 h at room temperature. Primary antibodies were as follows: anti-CXCL16 ab101404, 1:2000; anti-p65 ab16502, 1:2000; p-p65 antibody ab86299, 1:2000; anti-Rel-B ab154957, 1:5000; p-Rel-B antibody ab47366, 1:1000; anti-MMP1 ab38929, 1:5000; anti-MMP9 ab38898, 1:1000; anti-MMP13 ab39012, 1:6000; anti-GAPDH ab9485, 1:2500, and secondary antibody was Goat Anti-Rabbit IgG H&L (HRP) ab6721, 1:3000 (Abcam, Cambridge, MA, USA). GAPDH was used as an internal control. Visualization of chemo-luminescent signals was performed by BeyoECL Plus Kit (Beyotime, Shanghai, China). The intensity of each band was analyzed using Gel Pro Analyzer Software 4.0 (Media Cybernetics, Inc., Rockville, MD, USA).
Cell Proliferation Analysis
Cell Counting Kit-8 (CCK8; Solarbio, Beijing, China) assay was conducted to assess cell proliferation. Transfected TPC-1 and K-1 cells were seeded into 96-well plates at the density of 1×104 cells/well. After incubation for 0, 12, 24 and 48 h, respectively, 10 μL CCK-8 solution was added. Another incubation for 2 h, the value of optical density was recorded at 450 nm using a microplate reader (Multiscan FC, Thermo Fisher Scientific, Inc.).
Wound Healing Assay
Wound healing assay was performed to measure the capability of cell migration. Confluent transfected TPC-1 and K-1 monolayer cells were wounded using a 10 μL sterile pipette tip in 6-well plates. After washing with PBS, the cells were incubated for 24 h. Cell images were obtained with an inverted microscope (Olympus, Tokyo, Japan) and the distance between the wound sides was quantified.
Transwell Assay
For the cell invasion assay, 1×105 TPC-1 and K-1 cells suspended in 200 μL serum-free medium were plated in the upper chambers with Matrigel-coated membranes (24-well, 8μm pore size, BD, Franklin Lakes, NJ, USA). The lower chambers were filled with DMEM containing 10% FBS. After 24 h of incubation, non-invaded cells were removed by sterile swabs and invaded cells were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 15 min. The stained cells were imaged under a microscope (Olympus Corp, Tokyo, Japan), and were counted in five random visual fields.
Statistical Analysis
The study results were analyzed by GraphPad Prism 6.00 (GraphPad Software, San Diego, CA, USA). Data were presented as mean ± standard error of estimate (SEM) of at least three independent experiments. Paired Student’s test was used for comparisons of two groups and One-way ANOVA followed by Newman‑Keuls post hoc analysis was used for comparisons of multiple groups. Differences were considered statistically significant when P<0.05.
Results
miR-873-5p Is Expressed at Low Expression Levels in PTC Tissues and Cell Lines
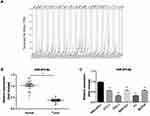
First, we performed the bioinformatic analysis by searching for miR-873-5p expression data in the public database. Figure 1A shows the expression of miR-873-5p in 33 different types of cancers, and it has been observed that miR-873-5p was significantly down-regulated in PTC tumor tissue (THCA) compared with the normal tissue. To determine the expression of miR-873-5p in PTC, RT-qPCR was performed to measure its expression in 30 pairs of PTC tissues and the adjacent normal tissues. As illustrated in Figure 1B, miR-873-5p expression was lower in tumor tissues than that in adjacent non-tumor tissues (P<0.01). In addition, the expression of miR-873-5p was also downregulated in PTC cell lines, including KTC-1, TPC-1, BHP10-3, K1 and BCPAP, compared with normal thyroid epithelial Nthy-ori3-1 cells (P<0.01; Figure 1C).
CXCL16 Is a Target of miR-873-5p
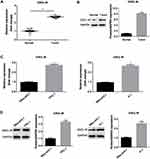
To investigate the target gene of miR-873-5p, we searched TargetScan online tool and CXCL16 was chosen as a target for further study (Figure 2A). Moreover, the results of the bioinformatic analysis also suggest the potential function of miR-873-5p in regulation of CXCL16 (Figure 2B). Then, dual-luciferase reporter assay was performed to confirm the targeted relationship. By co-transfection of pmirGLO-CXCL16-WT or pmirGLO-CXCL16-mutant together with miR-873-5p mimics or miR-NC mimics, the relative luciferase activity was measured. miR-873-5p decreased the relative luciferase activity in mutant 3ʹUTR of CXCL16, compared with miR-NC mimics (P<0.01; Figure 2C). However, there were no significant changes between miR-873-5p mimics and miR-NC mimics in WT CXCL16 3ʹUTR. These results indicated that CXCL16 is a target of miR-873-5p.
CXCL16 Expression Is Upregulated in PTC Tissues and Cells
Then, we detected the expression of CXCL16 in PTC tissues and adjacent normal tissues at both mRNA and protein levels by RT-qPCR and Western blot, respectively. The results showed that CXCL16 expression was higher in tumor tissue than that in matched adjacent normal tissues (P<0.01; Figure 3A and B). Moreover, CXCL16 expression was increased in TPC-1 and K1 cells at both mRNA and protein levels, compared with normal cell Nthy-ori3-1 (P<0.01; Figure 3C and D).
Overexpression of miR-873-5p Inhibits PTC Cell Proliferation, Migration and Invasion Through Targeting CXCL16
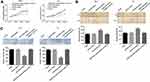
To evaluate cell proliferation, migration and invasion, TPC-1 and K-1 cells were transfected with miR-873-5p mimics and pcDNA3.1-CXCL16, and CCK-8, wound healing assay and transwell assay were performed in vitro. miR-873-5p mimics significantly increased the level of miR-873-5p in both TPC-1 and K-1 cells (p<0.01), and the transfection efficiency was shown in Figure S1. The data indicated that overexpression of miR-873-5p decreased cell proliferation, retarded wound healing and reduced the number of invaded cells in both TPC-1 and K-1 cells, while CXCL16 reversed the inhibition induced by miR-873-5p (P<0.01, Figure 4A–C). These results suggested that miR-873-5p suppressed the proliferation, migration and invasion of PTC cells via regulating CXCL16 expression.
miR-873-5p Suppresses the NF-κB Pathway and MMP1, MMP9, MMP13 Expression in PTC Cells by Targeting CXCL16
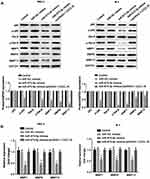
Finally, we investigated the effects of miR-873-5p and CXCL16 on NF-κB pathway and several MMPs expression. Overexpression of miR-873-5p significantly decreased the phosphorylation of p65 and Rel-B, while CXCL16 rescued the reduction induced by miR-873-5p in both TPC-1 and K-1 cells (Figure 5A). However, miR-873-5p and CXCL16 did not affect p65 and Rel-B protein levels. Additionally, miR-873-5p overexpressing repressed the mRNA and protein expressions of MMP1, MMP9 and MMP13, while CXCL16 abolished these repressions in TPC-1 and K-1 cells (P<0.01; Figure 5A and B). Taken together, these results demonstrated that miR-873-5p targets CXCL16 to suppress the NF-κB signaling pathway and MMP1, MMP9, MMP13 expression in PTC cells.
Discussion
In the present study, it was revealed that miR-873-5p was downregulated in PTC tissues and cell lines. CXCL16, a target of miR-873-5p, was upregulated in tumor tissues and PTC cells. Moreover, overexpression of miR-873-5p has the inhibition effect on the proliferation, migration and invasion by targeting CXCL16 in PTC cells.
Accumulating evidence revealed that miR-873 functions as a tumor suppressor in several human cancers. For instance, overexpression of miR-873 reduces glioblastoma multiforme cell proliferation, migration and invasion through regulating IGF2BP1 expression.13 Moreover, miR-873-5p expresses low in colorectal cancer, and its overexpression inhibits cell migration, invasion and epithelial–mesenchymal transition (EMT) by targeting ZEB1.14 However, there are also studies showed that miR-873 functions as an oncogene. For example, miR-873 promotes the proliferation and migration in lung adenocarcinoma cells.15 Besides, in hepatocellular carcinoma, miR-873 is upregulated in tissues and cells, and knockdown of which inhibits cell growth and metastasis.16 In addition, research about miR-873 have focused on drug resistance, such as mediating cisplatin and paclitaxel combination resistance in ovarian cancer17 and regulating gefitinib resistance in non-small cell lung cancer.18 In our study, we explored the biological roles of miR-873-5p in PTC. The expression of miR-873-5p was upregulated in PTC tissues and cell lines. Moreover, overexpression of miR-873-5p inhibits PTC cell proliferation, migration and invasion. These results suggested that miR-873-5p has tumor-suppressive effects on PTC, consistent with its role in colorectal cancer.14
CXCL16 is a member of the CXC chemokine family, and the structure of CXCL16 is similar to fractalkine (neurotactin) in having a transmembrane region and a chemokine domain.19 The interaction between chemokines and chemokine receptors is vital in the process of cancer metastasis in most aggressive cancers, such as CXCL16 and its receptor CXCR6.20 CXCL16 is aberrantly expressed in various cancers and could regulate cell proliferation, migration and invasion of the cancer cells. For example, in hepatocellular carcinoma, CXCL16 is highly expressed in cancer-associated fibroblasts than peri-tumor fibroblasts, and it promotes cell migration and invasion.21 Additionally, overexpression of CXCL16 promotes proliferation and migration of schwannomas.22 Furthermore, reduction of CXCL16 inhibits the viability and invasiveness in lung cancer cell lines.23 In PTC, a previous study has reported that treatment with CXCL16 enhances macrophage-mediated PTC cell migration, and blocking CXCL16 signaling attenuates the enhancement.24 However, the biological roles of CXCL16 in PTC remain largely unknown. In the present study, CXCL16 was identified as a target of miR-873-5p. The expression of CXCL16 was increased in PTC tissues and cells. Moreover, CXCL16 could reverse the inhibition on cell proliferation, migration and invasion induced by miR-873-5p. These results suggested that miR-873-5p inhibits the proliferation, migration and invasion in PTC cells by targeting CXCL16.
NF-κB signaling pathway is activated in various types of cancers, which could promote tumorigenesis.25,26 P65 and Rel-B are two critical members of the NF-κB family. It was reported that NF-κB activation is a benefit to PTC tumor growth and aggressiveness.27 Furthermore, a previous study revealed that silencing of CXCL16 suppressed proliferation and invasion of lung cancer cells through regulating NF-κB pathway.28 MMPs are associated with the development of caners, through which extracellular matrix degradation, and cell invasion and metastasis were enhanced.29,30 Additionally, in the early stage of tumorigenesis, MMPs may also modulate angiogenesis.31 In this study, we explored the effects of miR-873-5p and CXCL16 on NF-κB pathway and several MMPs (MMP1, MMP9 and MMP13) expression. Overexpression of miR-873-5p suppressed the NF-κB pathway and MMP1, MMP9 and MMP13 expression in PTC cells, while CXCL16 reversed this suppression. The data suggested that miR-873-5p inhibits PTC cell proliferation, migration and invasion by targeting CXCL16 through inactivating the NF-κB signaling pathway and repressing MMP1, MMP9 and MMP13 expression.
Conclusion
MiR-873-5p functions as a tumor suppressor in PTC, andCXCL16 is identified as a target of miR-873-5p. Importantly, overexpression of miR-873-5p inhibits the proliferation, migration and invasion through suppressing NF-κB signaling pathway and MMP1, MMP9, MMP13 expression by targeting CXCL16 in PTC cells. These findings suggest that miR-873-5p might be a potential new target for PTC.
Abbreviations
CCK8, Cell Counting Kit-8; CXCL16, C-X-C motif chemokine ligand 16; miRNA, microRNA; MMP, matrix metalloprotein; NC, negative control; PTC, papillary thyroid cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; UTR, untranslated region; WT, wild type.
Funding
This study was supported by the Key Clinical Discipline in Shanghai-Department of General Surgery (No. 2017ZZ02007).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Xie J, Fan Y, Zhang X. Molecular mechanisms in differentiated thyroid cancer. Front Biosci (Landmark Ed). 2016;21:119–129. doi:10.2741/4379
2. Radu TG, Ciurea ME, Mogoantă SŞ, et al. Papillary thyroid cancer stroma - histological and immunohistochemical study. Rom J Morphol Embryol. 2016;57(2 Suppl):801–809.
3. Lubitz CC, Sosa JA. The changing landscape of papillary thyroid cancer: epidemiology, management, and the implications for patients. Cancer. 2016;122:3754–3759. doi:10.1002/cncr.v122.24
4. Mutalib N-SA, Yusof AM, Mokhtar NM, et al. MicroRNAs and lymph node metastasis in papillary thyroid cancers. Asian Pac J Cancer Prev. 2016;17:25–35. doi:10.7314/APJCP.2016.17.1.25
5. Qiu Z, Li H, Wang J, et al. miR-146a and miR-146b in the diagnosis and prognosis of papillary thyroid carcinoma. Oncol Rep. 2017;38:2735–2740. doi:10.3892/or.2017.5994
6. Miccoli P, Bakkar S. Surgical management of papillary thyroid carcinoma: an overview. Updates Surg. 2017;69:145–150. doi:10.1007/s13304-017-0449-5
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi:10.1016/S0092-8674(04)00045-5
8. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
9. Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi:10.1016/j.molmed.2014.06.005
10. Lee JC, Gundara JS, Glover A, Serpell J, Sidhu SB. MicroRNA expression profiles in the management of papillary thyroid cancer. Oncologist. 2014;19:1141–1147. doi:10.1634/theoncologist.2014-0135
11. Saiselet M, Gacquer D, Spinette A, et al. New global analysis of the microRNA transcriptome of primary tumors and lymph node metastases of papillary thyroid cancer. BMC Genomics. 2015;16:828. doi:10.1186/s12864-015-2082-3
12. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods. 2001;25:402–408. doi:10.1006/meth.2001.1262
13. Wang RJ, Li JW, Bao BH, et al. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938–8948. doi:10.1074/jbc.M114.624700
14. Li G, Xu Y, Wang S, et al. MiR-873-5p inhibits cell migration, invasion and epithelial-mesenchymal transition in colorectal cancer via targeting ZEB1. Pathol Res Pract. 2019;215:34–39. doi:10.1016/j.prp.2018.10.008
15. Gao Y, Xue Q, Wang D, et al. miR-873 induces lung adenocarcinoma cell proliferation and migration by targeting SRCIN1. Am J Transl Res. 2015;7:2519–2526.
16. Han G, Zhang L, Ni X, et al. MicroRNA-873 promotes cell proliferation, migration, and invasion by directly targeting TSLC1 in hepatocellular carcinoma. Cell Physiol Biochem. 2018;46:2261–2270. doi:10.1159/000489594
17. Wu DD, Li XS, Meng XN, et al. MicroRNA-873 mediates multidrug resistance in ovarian cancer cells by targeting ABCB1. Tumour Biol. 2016;37:10499–10506. doi:10.1007/s13277-016-4944-y
18. Jin S, He J, Li J, et al. MiR-873 inhibition enhances gefitinib resistance in non-small cell lung cancer cells by targeting glioma-associated oncogene homolog 1. Thorac Cancer. 2018;9:1262–1270. doi:10.1111/tca.2018.9.issue-10
19. Wilbanks A, Zondlo SC, Murphy K, et al. Expression cloning of the STRL33/BONZO/TYMSTRligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol. 2001;166:5145–5154. doi:10.4049/jimmunol.166.8.5145
20. Deng L, Chen N, Li Y, et al. CXCR6/CXCL16 functions as a regulator in metastasis and progression of cancer. Biochim Biophys Acta. 2010;1806:42–49. doi:10.1016/j.bbcan.2010.01.004
21. Liu J, Chen S, Wang W, et al. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-β pathways. Cancer Lett. 2016;379:49–59. doi:10.1016/j.canlet.2016.05.022
22. Held-Feindt J, Rehmke B, Mentlein R, et al. Overexpression of CXCL16 and its receptor CXCR6/bonzo promotes growth of human schwannomas. Glia. 2008;56:764–774. doi:10.1002/(ISSN)1098-1136
23. Hu W, Liu Y, Zhou W, et al. CXCL16 and CXCR6 are coexpressed in human lung cancer in vivo and mediate the invasion of lung cancer cell lines in vitro. PLoS One. 2014;9:e99056. doi:10.1371/journal.pone.0099056
24. Cho SW, Kim YA, Sun HJ, et al. CXCL16 signaling mediated macrophage effects on tumor invasion of papillary thyroid carcinoma. Endocr Relat Cancer. 2016;23:113–124. doi:10.1530/ERC-15-0196
25. Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi:10.1186/1476-4598-12-86
26. Xia Y, Shen S, Verma IM. NF-κB, an active player in human cancers. Cancer Immunol Res. 2014;2:823–830. doi:10.1158/2326-6066.CIR-14-0112
27. Pyo JS, Kang G, Kim DH, et al. Activation of nuclear factor-κB contributes to growth and aggressiveness of papillary thyroid carcinoma. Pathol Res Pract. 2013;209:228–232. doi:10.1016/j.prp.2013.02.004
28. Liang K, Liu Y, Eer D, et al. High CXC chemokine ligand 16 (CXCL16) expression promotes proliferation and metastasis of lung cancer via regulating the NF-κB pathway. Med Sci Monit. 2018;24:405–411. doi:10.12659/MSM.906230
29. Shay G, Lynch CC, Fingleton B. Moving targets: emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2015;44–46:200–206. doi:10.1016/j.matbio.2015.01.019
30. Kessenbrock K, Wang CY, Werb Z. Matrix metalloproteinases in stem cell regulation and cancer. Matrix Biol. 2015;44–46:184–190. doi:10.1016/j.matbio.2015.01.022
31. Folgueras AR, Pendás AM, Sánchez LM, et al. Matrix metalloproteinases in cancer: from new functions to improved inhibition strategies. Int J Dev Biol. 2004;48:411–424. doi:10.1387/ijdb.041811af
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.