Back to Journals » OncoTargets and Therapy » Volume 12
miR-7 Reverses Breast Cancer Resistance To Chemotherapy By Targeting MRP1 And BCL2
Received 28 April 2019
Accepted for publication 7 October 2019
Published 16 December 2019 Volume 2019:12 Pages 11097—11105
DOI https://doi.org/10.2147/OTT.S213780
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Tianzi Hong,1 Jian Ding,2 Wenlian Li2
1Department of Thyroid and Breast Surgery, Jinjiang Hospital of Quanzhou Medical College, Jinjiang 362200, People’s Republic of China; 2Department of Breast Surgery, Zhongshan Hospital of Xiamen University, Xiamen 361004, People’s Republic of China
Correspondence: Wenlian Li
Department of Breast Surgery, Zhongshan Hospital of Xiamen University, No. 201 South Hubin Street, Siming District, Xiamen 361004, People’s Republic of China
Tel +86 0592-2993152
Email [email protected]
Background: MicroRNAs (miRNAs) are a class of non‐coding RNAs that have been linked with breast cancer chemoresistance, which is a major clinical problem causing disease relapse and poor prognosis. miR-7 exerts several tumor suppressive activities.
Purpose: This study was designed to clarify whether and how miR-7 regulates breast cancer chemoresistance.
Methods: miR-7 level in breast cancer was determined by qRT-PCR analysis. Cell viability was assessed by MTS assay to quantify the IC50 value of paclitaxel and carboplatin. The targets of miR-7 were confirmed by luciferase reporter assay.
Results: Higher miR-7 expression predicts better pathological complete response (pCR) of breast cancer patients receiving paclitaxel/carboplatin chemotherapy. In vitro, miR-7 sensitizes breast cancer cell lines (MCF-7 and MDA-MB-231) to paclitaxel and carboplatin, alone and in combination. In addition, we reveal that both the multidrug resistance-associated protein 1 (MRP1) and anti-apoptotic B cell lymphoma 2 (BCL2) are targets of miR-7 in breast cancer cells. Furthermore, miR-7-induced sensitization of breast cancer to paclitaxel/carboplatin is markedly reversed by restoration of MRP1 and BCL2.
Conclusion: These findings show that miR-7 reverses breast cancer chemoresistance through suppressing MRP1 and BCL2, and also suggest that miR-7 may possess a predictive value and represent a therapeutic target in breast cancer chemotherapy.
Keywords: miR-7, breast cancer, pathological complete response, chemoresistance, MRP1, BCL2
Introduction
Breast cancer is the most common malignancy and the leading cause of cancer-related death in women.1 Owing to the development of modern mammographic screening and neoadjuvant chemotherapy, the prognosis of breast cancer patients has been largely improved over the past decades.2,3 However, despite different treatment modalities, chemoresistance frequently emerges in breast cancer patients,4 which remains to be a major clinical impediment to achieve successful treatment.5 To discover more effective therapeutics, a better understanding of the molecular mechanisms of breast cancer chemoresistance is warranted.
In addition to its effectiveness and increasing usage for treating newly diagnosed or locally advanced breast cancer, neoadjuvant chemotherapy also permits the assessment of treatment response, which holds the potentiality to avoid ineffective regimens and help to discover novel therapeutic targets.6 The pathologic complete response (pCR) has recently been used as a prognostic parameter associated with improved long-term outcomes following neoadjuvant chemotherapy.7 Therefore, searching novel biomarkers for predicting pCR would be clinically valuable for guiding the utilization of neoadjuvant chemotherapy.
microRNAs (miRNAs) are short noncoding RNA molecules that negatively regulate gene expression via targeting mRNAs.8 In recent years, increasing evidence has shown that some miRNAs, such as miR-6219 and miR-205,10 are involved in chemoresistance regulation and also possess predictive value for chemotherapy response of breast cancer, implicating that miRNAs could serve as promising predictive biomarkers and therapeutic targets in breast cancer treatment. Studies have reported that miR-7 inhibits the invasion and metastasis of breast cancer.11,12 Lately, the intratumoral level of miR-7 was shown to be linked with breast cancer response to anthracycline/taxane-based neoadjuvant chemotherapy.13 However, whether miR-7 predicts pCR to other regimens needs further investigation, and secondly, whether it regulates chemoresistance in breast cancer is unknown either. In this study, we investigated the predictive value of miR-7 in pCR following paclitaxel plus carboplatin chemotherapy and explored the mechanisms by which miR-7 regulates breast cancer chemoresistance.
Materials And Methods
Patient Samples
A total of 60 breast cancer patients manifesting pathological complete response (pCR, n = 28) or not (non-pCR, n = 32) following 4 cycles of paclitaxel plus carboplatin chemotherapy (paclitaxel, 80 mg/m2; carboplatin at an area under the curve of 2 mg × min/mL) were recruited in this study (Figure 1). The study was approved by the ethical committee of Jinjiang Hospital of Quanzhou Medical College. The written informed consent was obtained from all patients. Patients without invasive tumors in the final surgical breast and axillary lymph nodes or with residual ductal carcinoma in situ were included into pCR group. Breast cancer tissues were obtained by a core biopsy before chemotherapy and stored immediately at −80°C.
Cell Lines And Viability Measurement
Human breast cancer cell lines MCF-7 and MDA-MB-231, and HEK293 cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The paclitaxel-resistant MCF-7 cells (MCF-7-PR) were established and maintained in culture medium with 10 μg/mL paclitaxel. All cells were cultured under standard conditions. MCF-7, MCF-7-PR, and MDA-MB-231 cells were treated with serial dilutions of paclitaxel (1–100 μg/mL) (Sigma) and carboplatin (0.05–3 mg/mL) (Sigma) for 72 hrs. Cell viability was determined by MTS assay using the CellTiter 96 AQueous Solution Reagent kit (Promega) according to the manufacturer’s instructions. IC50 values were calculated. Each treatment was carried out with 5 replicates.
Lentiviral Infection
Human MRP1 gene and BCL2 gene were amplified by PCR and individually cloned into the pCDH-CMV-Puro lentiviral vector (System Biosciences, Palo Alto, CA, USA). Lentivirus was produced as previously described.14 MCF-7 and MDA-MB-231 cells were infected with vector control, MRP1-overexpressing, or BCL2-overexpressing lentivirus at a multiplicity of infection (MOI) of 100 in the presence of 10 µg/mL polybrene. The stably infected cells were selected by puromycin. The overexpression was confirmed by Western blotting analysis as described below.
Western Blotting
Total protein extracts were obtained through the homogenization of cells in RIPA lysis buffer on ice for 20 mins, followed by centrifugation at 10,000 × g for 10 mins at 4°C. Equal amount of proteins were separated by SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were probed with primary antibodies against MRP1 (Abcam, ab24102), BCL2 (Proteintech, 12789-1-AP), and β-Actin (Santa Cruz, sc-47778). Then, membranes were incubated with HRP-conjugated secondary antibodies (Santa Cruz) and protein bands were developed by the ECL Western blotting detection system (GE Healthcare). Protein bands were quantified using the ImageJ 1.63 (National Institutes of Health, Bethesda, MD, USA).
qRT-PCR Analysis
Total RNA was isolated with Trizol reagent (Invitrogen) and reverted to cDNA using the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher Scientific) according to the manufacturers’ instructions. qRT-PCR reactions were performed in triplicates using synthetic primers (RiboBio), SYBR Green qPCR Supermix (Invitrogen), and CFX96 real-time PCR System (Bio-Rad). Primers were available upon request. Cycle threshold (Ct) values were determined, and expression levels were normalized to U6 or β-Actin in each reaction.
Cell Transfection And Luciferase Reporter Assay
The control mimic, miR-7 mimic, control inhibitor, and miR-7 inhibitor were purchased from GenePharma (Shanghai, China) and transfected into MCF-7 and MDA-MB-231 cells using the Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocols (final concentration: 100 nM). At 48 hrs after transfection, cells were collected for measuring miR-7 expression or used for further experiments. For luciferase reporter assay, 3ʹ-UTR fragment of MRP1 and BCL2 was cloned into the pGL3-control vectors (Promega). The mutant construct was developed by using the Phusion Site-Directed Mutagenesis Kit (ThermoFisher Scientific). HEK293 cells were seeded in 24-well plates and co-transfected firefly luciferase reporter vector, control Renilla pRL-TK vector (Promega) with control mimic, miR-7 mimic, control inhibitor, or miR-7 inhibitor using the Lipofectamine 2000. At 48 hrs after transfection, cells were lysed and the activity of firefly and Renilla luciferase was assessed using the Dual-luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Each treatment was performed with 4 replicates, and firefly luciferase activity was normalized to Renilla luciferase activity for each well.
Statistical Analysis
Statistical analysis was performed using the SPSS 24.0 (SPSS Inc, Chicago, IL, USA). Patient disease-free survival (DFS) was analyzed by the Kaplan–Meier method, and miR-7 predictive value was analyzed by ROC curve. Data were expressed as the mean ± s.e.m., and compared by two-tailed Student’s t-test or one-way ANOVA analysis. P value < 0.05 was considered statistically significant for all analyses.
Results
miR-7 Level Predicts Pathological Complete Response Of Breast Cancer Patients
Paclitaxel plus carboplatin regimen has long been used as a first-line neoadjuvant chemotherapy for women with metastatic breast cancer.15,16 However, reliable clinical parameters capable of predicting therapeutic response to this regimen are unavailable. To examine whether miR-7 has predictive value for pathological complete response (pCR), a prognostic parameter defined by absence of invasive cancer following treatment,7 we compared its intratumoral expression between breast cancer patients achieved pCR (pCR, n = 28) and those did not achieve pCR (non-pCR, n = 32) following 4 cycles of paclitaxel plus carboplatin chemotherapy. Patients’ clinical information and demographics including the expression of miR-7 are summarized in Figure 1A. Quantitative real-time PCR (qRT-PCR) analysis showed that miR-7 level was significantly higher in pCR patients than that in non-pCR patients (Figure 1B, p < 0.05), suggesting that higher miR-7 level is associated with a better therapeutic response to paclitaxel plus carboplatin regimen in breast cancer patients. Patients who achieved pCR in this analysis are currently disease-free, and the Kaplan–Meier analysis showed that pCR was correlated with an improved DFS (data not shown). We next investigated whether miR-7 level is associated with patient’s DFS. To address it, the whole 60 breast cancer patients were equally stratified into miR-7-low and miR-7-high groups. The Kaplan–Meier analysis revealed that patients in miR-7-high group had a significantly longer DFS than those in miR-7-low group (Figure 1C, p < 0.05). Therefore, these results suggest that higher miR-7 level predicts a better pCR to paclitaxel plus carboplatin chemotherapy, which is associated with a favorable long-term outcome in breast cancer patients.
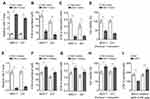
miR-7 Sensitizes Breast Cancer To Paclitaxel And Carboplatin
The earlier studies on clinical samples suggest that higher miR-7 level is associated with higher sensitivity to paclitaxel plus carboplatin chemotherapy. Next, to gain insight into the functional role of miR-7 in breast cancer chemoresistance, synthetic mimic was transfected into two human breast cancer cell lines, MCF-7 and MDA-MB-231, for overexpressing miR-7. The efficient enforced expression of miR-7 in both cell lines was confirmed by qRT-PCR analysis, as compared with negative control (NC) mimic (Figure 2A). Next, these cells were treated with serial dilutions of paclitaxel and carboplatin, and cell viability was then measured by MTS assay. Results showed that in contrast to NC mimic transfection, the IC50 values of paclitaxel (Figure 2B) and carboplatin (Figure 2C) were drastically lower in both MCF-7 and MDA-MB-231 cells overexpressed with miR-7. Similarly, cell viability of miR-7-overexpressing MCF-7 and MDA-MB-231 (Figure 2D) cells was overtly decreased in response to paclitaxel plus carboplatin, indicating that these cells display increased sensitivity to both paclitaxel and carboplatin. To strengthen these observations, miR-7 was knocked down in MCF-7 and MDA-MB-231 cells via transfecting miR-7 inhibitor, which was validated by qRT-PCR analysis (Figure 2E). Notably, in agreement with miR-7 overexpression, its knockdown resulted in higher IC50 values of paclitaxel (Figure 2F) and carboplatin (Figure 2G), and elevated cell viability (Figure 2H) in MCF-7 and MDA-MB-231 cells treated with paclitaxel and carboplatin. Furthermore, consistently, miR-7 overexpression drastically reduced paclitaxel IC50 value, and in reverse, miR-7 inhibition increased it in paclitaxel-resistant MCF-7 cells (MCF-7-PR) (Figure 2I). Overall, these data show that miR-7 reveres chemoresistance in breast cancer cells, at least in vitro.
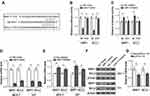
MRP1 And BCL2 Are Both Targets Of miR-7 In Breast Cancer
To elucidate how miR-7 regulates breast cancer chemoresistance, its potential mRNA targets were predicted by in-silico algorithms (TargetScan and miRBase).17 Among these putative targets (Supplementary Table 1), we found two chemoresistance-related molecules, including the multiple drug resistance protein 1 (MRP1)18 and B-cell lymphoma 2 (BCL2)19 (Figure 3A). As shown by luciferase reporter assay, miR-7 overexpression inhibited and its knockdown increased luciferase activity of both MRP1 and BCL2 wild-type constructs, meanwhile, with mutant ones unaffected (Figure 3B-C). Moreover, in MCF-7 and MDA-MB-231 cells, miR-7 overexpression suppressed and its knockdown increased mRNA level (Figure 3D–E) and protein level (Figure 3F) of MRP1 and BCL2. Furthermore, contrary to a significant higher miR-7 expression in pCR patients (Figure 1B), the mRNA levels of MRP1 and BCL2 were both found downregulated in these patients (Figure 3G), thus further proving that MRP1 and BCL2 are direct targets of miR-7 in breast cancer.
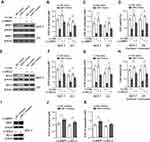
Targeted MRP1 And BCL2 Both Contribute To miR-7-Induced Sensitization Of Breast Cancer To Paclitaxel And Carboplatin
To understand whether MRP1 and BCL2 are involved in miR-7 regulation of breast cancer response to paclitaxel and carboplatin, the expression of MRP1 (Figure 4A) and BCL2 (Figure 4E) was restored by lentiviral infection in miR-7-overexpressing MCF-7 and MDA-MB-231 cells. Evidently, along with MRP1 restoration, miR-7-decreased IC50 values of paclitaxel (Figure 4B) and carboplatin (Figure 4C) in both MCF-7 and MDA-MB-231 cells were largely rescued. Moreover, MRP1 restoration also led to significant recovery in cell viability when treated with paclitaxel plus carboplatin (Figure 4D). These findings indicate that the suppressed MRP1 contributes to miR-7-increased breast cancer sensitivity to paclitaxel and carboplatin. Alternatively, similar to MRP1, the restoration of BCL2 expression also prominently rescued IC50 values of paclitaxel (Figure 4F) and carboplatin (Figure 4G), as well as cell viability (Figure 4H), in MCF-7 and MDA-MB-231 cells exposed to paclitaxel and carboplatin treatment. Moreover, in contrast to miR-7 overexpression, its inhibition resulted in significant increase in IC50 values of paclitaxel (Figure 4J) and carboplatin (Figure 4K) in MCF-7 cells overexpressed with both MRP1 and BCL-2 (Figure 4I). Altogether, according to these mechanistic evidence, we propose that miR-7 increases breast cancer sensitivity to paclitaxel and carboplatin through inhibiting both MRP1 and BCL2.
Discussion
Despite great improvement in breast cancer treatment, the frequently emerging chemoresistance remains a major challenge for improving the effectiveness of chemotherapeutic agents and clinical outcome of breast cancer patients.20 Owing to several advantages compared with traditional chemotherapy, including the permission of evaluating tumor response to treatment, neoadjuvant chemotherapy, a type of chemotherapy before surgical treatment, is commonly used as a standard option for treating locally advanced breast cancer.21,22 pCR is a pathological parameter defined as a complete clinical response with no residual invasive disease when evaluating the surgical breast specimen in women.23 Studies have shown that pCR achievement after neoadjuvant treatment is strongly associated with a better long-term outcome,24,25 for which, albeit, pCR is still not validated as a surrogate endpoint.7 In our study, we show that higher miR-7 is a favorable biomarker for predicting better pCR in breast cancer patients receiving paclitaxel plus carboplatin neoadjuvant chemotherapy. In addition to the investigations on clinical samples, we subsequently demonstrate that miR-7 sensitizes breast cancer cells to these two agents at least by targeting two key chemoresistance-associated molecules, MRP1 and BCL2, therefore establishing a potential molecular mechanistic link between miR-7-conferred better response to paclitaxel plus carboplatin neoadjuvant chemotherapy and reduced chemoresistance. In this sense, our findings may shed new light on miR-7 regulation of breast cancer chemoresistance.
The expression profile of miRNAs has been associated with pathologic response to neoadjuvant chemotherapy.26 In a recent study, intratumoral miR-7 expression was reported to predict pCR in breast cancer patients treated with anthracycline/taxane-based neoadjuvant chemotherapy.13 In this study, we show that miR-7 also has a predictive value for pCR and higher miR-7 level is significantly associated with longer DFS in patients received paclitaxel plus carboplatin treatment; thus, these observations may extend its potential application to predict response to another specific regimen in breast cancer patients. Our results may also highlight miR-7 as a potential biomarker to guide the neoadjuvant chemotherapy in breast cancer patients in the future. However, it should be noted that the clinical sample size is relatively limited. Further analogous studies would be required to consolidate the predictive value of miR-7 in pCR. Moreover, to our knowledge, miR-7 has also been associated with prognosis of patients with colorectal cancer27 and lung cancer.28 We speculate that miR-7 could also be useful for predicting pCR in these cancers. It is of interest and clinical significance to examine whether this is the case in future investigations.
Functionally, several previous studies have associated miR-7 roles with breast cancer pathologies, mostly documenting its tumor suppressive effects. For instance, miR-7 inhibits the epithelial–mesenchymal transition and metastasis of breast cancer stem cells by downregulating the STAT3 pathway29 and the Kruppel-like factor 4.30 miR-7 also suppresses the proliferation and induces apoptosis of breast cancer cells via targeting the proteasome activator subunit 3γ.31 Although miR-7 has been connected to cisplatin and adriamycin resistance in breast cancer cells, in which the modulated REGγ32 and EGFR/PI3K signaling pathway33 play a role, it remains unclear whether and how it is associated with the resistance of paclitaxel and carboplatin, two chemotherapeutic agents received by breast cancer patients enrolled in our study. By utilizing in vitro experimental system wherein cultured MCF-7 and MDA-MB-231 were treated with paclitaxel and carboplatin alone or in combination, we reveal that miR-7 acts as a suppressor in chemoresistance to both of agents. Furthermore, we also obtained similar results through investigating a paclitaxel-resistant MCF-7 cell line, consolidating our conclusion on miR-7 negative regulation of chemoresistance to paclitaxel and carboplatin in breast cancer cells. Together with the previous reports, we guess that miR-7 may function to reverse breast cancer resistance to a broader range of chemotherapeutic agents.
To date, multiple molecular mechanisms have been associated with chemoresistance in cancer, such as upregulation of transporter pumps, anti-apoptosis, and dysregulation of miRNAs.34,35 MRP1 mediates ATP-dependent efflux of drugs from cells and its elevated expression predicts poor response to chemotherapy in several cancers.36 In addition, BCL2 has long been recognized to confer chemoresistance by negatively regulating apoptosis of cancer cells under cytotoxic conditions.19 We demonstrate that miR-7 sensitizes breast cancer to paclitaxel and carboplatin by targeting MRP1 and BCL2 since the restoration of both MRP1 and BCL2 rescues this effect of miR-7. More importantly, this mechanistic evidence obtained in vitro provides a molecular basis for explaining how higher miR-7 level is associated with better response of breast cancer to paclitaxel plus carboplatin neoadjuvant chemotherapy. One major limitation of this study is the lack of in vivo evidence. Further studies such as those using xenograft animal models would be helpful to validate the inhibitory effect of miR-7 on breast cancer chemoresistance. Another interesting topic needed to be elucidated in the future is whether the regulation of apoptosis, particularly through BCL2, is involved in the process during which miR-7 influences breast cancer chemoresistance. As we have shown in the list of Supplementary Table 1, there are nearly 6000 genes that are potential candidates of miR-7 targets. Despite the infeasibility to fully survey the involvement of each of them in mediating miR-7 function, which holds a great interest for further studies, our available data undoubtedly exemplify the miR-7/MRP1 and miR-7/BCL2 axes as two potential targets to reverse chemoresistance in breast cancer.
In conclusion, we reveal the potential of miR-7 as a new biomarker for predicting breast cancer response to chemotherapy and also mechanistically link miR-7-improved chemosensitivity with the inhibition of MRP1 and BCL2, which may provide a theoretical foundation for its utilization in counteracting chemoresistance and predicting patients’ response to neoadjuvant chemotherapy.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Plevritis SK, Munoz D, Kurian AW, et al. Association of screening and treatment with breast cancer mortality by molecular subtype in US women, 2000–2012. Jama. 2018;319(2):154–164. doi:10.1001/jama.2017.19130
3. Bleyer A, Welch HG. Effect of three decades of screening mammography on breast-cancer incidence. N Engl J Med. 2012;367(21):1998–2005. doi:10.1056/NEJMoa1206809
4. Tang Y, Wang Y, Kiani MF, Wang B. Classification, treatment strategy, and associated drug resistance in breast cancer. Clin Breast Cancer. 2016;16(5):335–343. doi:10.1016/j.clbc.2016.05.012
5. Gu G, Dustin D, Fuqua SA. Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Curr Opin Pharmacol. 2016;31:97–103. doi:10.1016/j.coph.2016.11.005
6. Cain H, Macpherson IR, Beresford M, Pinder SE, Pong J, Dixon JM. Neoadjuvant therapy in early breast cancer: treatment considerations and common debates in practice. Clin Oncol. 2017;29(10):642–652. doi:10.1016/j.clon.2017.06.003
7. Cortazar P, Geyer CE
8. Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
9. Xue J, Chi Y, Chen Y, et al. MiRNA-621 sensitizes breast cancer to chemotherapy by suppressing FBXO11 and enhancing p53 activity. Oncogene. 2016;35(4):448–458. doi:10.1038/onc.2015.96
10. Hu Y, Qiu Y, Yague E, Ji W, Liu J, Zhang J. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016;7(6):e2291. doi:10.1038/cddis.2016.194
11. Kong X, Li G, Yuan Y, et al. MicroRNA-7 inhibits epithelial-to-mesenchymal transition and metastasis of breast cancer cells via targeting FAK expression. PLoS ONE. 2012;7(8):e41523. doi:10.1371/journal.pone.0041523
12. Yu N, Huangyang P, Yang X, et al. microRNA-7 suppresses the invasive potential of breast cancer cells and sensitizes cells to DNA damages by targeting histone methyltransferase SET8. J Biol Chem. 2013;288(27):19633–19642. doi:10.1074/jbc.M113.475657
13. Raychaudhuri M, Bronger H, Buchner T, Kiechle M, Weichert W, Avril S. MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2017;162(3):511–521. doi:10.1007/s10549-017-4132-9
14. Al Yacoub N, Romanowska M, Haritonova N, Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J Gene Med. 2007;9(7):579–584. doi:10.1002/(ISSN)1521-2254
15. Burris H
16. Vernieri C, Milano M, Mennitto A, et al. Antitumor activity and safety profile of weekly carboplatin plus paclitaxel in metastatic breast cancer: a ten-year, monocentric, retrospective study. Breast Cancer Res Treat. 2017;165(2):365–373. doi:10.1007/s10549-017-4336-z
17. Dweep H, Sticht C, Gretz N. In-Silico algorithms for the screening of possible microRNA binding sites and their interactions. Curr Genomics. 2013;14(2):127–136. doi:10.2174/1389202911314020005
18. Munoz M, Henderson M, Haber M, Norris M. Role of the MRP1/ABCC1 multidrug transporter protein in cancer. IUBMB Life. 2007;59(12):752–757. doi:10.1080/15216540701736285
19. Maji S, Panda S, Samal SK, et al. Bcl-2 antiapoptotic family proteins and chemoresistance in cancer. Adv Cancer Res. 2018;137:37–75.
20. Kim C, Gao R, Sei E, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879–893 e813. doi:10.1016/j.cell.2018.03.041
21. Mieog JS, van der Hage JA, van de Velde CJ. Neoadjuvant chemotherapy for operable breast cancer. Br J Surg. 2007;94(10):1189–1200. doi:10.1002/bjs.5894
22. Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov. 2014;4(2):232–245. doi:10.1158/2159-8290.CD-13-0286
23. Pennisi A, Kieber-Emmons T, Makhoul I, Hutchins L. Relevance of pathological complete response after neoadjuvant therapy for breast cancer. Breast Cancer: Basic and Clinical Research. 2016;10:103–106. doi:10.4137/BCBCR.S33163
24. Bear HD, Anderson S, Smith RE, et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer:national surgical adjuvant breast and bowel project protocol B-27. J Clin Oncol. 2006;24(13):2019–2027. doi:10.1200/JCO.2005.04.1665
25. Kong X, Moran MS, Zhang N, Haffty B, Yang Q. Meta-analysis confirms achieving pathological complete response after neoadjuvant chemotherapy predicts favourable prognosis for breast cancer patients. Eur J Cancer. 2011;47(14):2084–2090. doi:10.1016/j.ejca.2011.06.014
26. Kolacinska A, Morawiec J, Fendler W, et al. Association of microRNAs and pathologic response to preoperative chemotherapy in triple negative breast cancer: preliminary report. Mol Biol Rep. 2014;41(5):2851–2857. doi:10.1007/s11033-014-3140-7
27. Suto T, Yokobori T, Yajima R, et al. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis. 2015;36(3):338–345. doi:10.1093/carcin/bgu242
28. Zhao J, Wang K, Liao Z, et al. Promoter mutation of tumor suppressor microRNA-7 is associated with poor prognosis of lung cancer. Mol Clin Oncol. 2015;3(6):1329–1336. doi:10.3892/mco.2015.648
29. Zhang H, Cai K, Wang J, et al. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32(11):2858–2868. doi:10.1002/stem.1795
30. Okuda H, Xing F, Pandey PR, et al. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73(4):1434–1444. doi:10.1158/0008-5472.CAN-12-2037
31. Shi Y, Luo X, Li P, et al. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGgamma. Cancer Lett. 2015;358(1):27–36. doi:10.1016/j.canlet.2014.12.014
32. Yang W, Yang X, Wang X, et al. Silencing CDR1as enhances the sensitivity of breast cancer cells to drug resistance by acting as a miR-7 sponge to down-regulate REGγ. J Cell Mol Med. 2019;23(8):4921–4932. doi:10.1111/jcmm.2019.23.issue-8
33. Huang Q, Wu YY, Xing SJ, Yu ZW. Effect of miR-7 on resistance of breast cancer cells to adriamycin via regulating EGFR/PI3K signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(12):5285–5292. doi:10.26355/eurrev_201906_18195
34. Zheng HC. The molecular mechanisms of chemoresistance in cancers. Oncotarget. 2017;8(35):59950–59964. doi:10.18632/oncotarget.19048
35. Wang J, Yang M, Li Y, Han B. The role of MicroRNAs in the chemoresistance of breast cancer. Drug Dev Res. 2015;76(7):368–374. doi:10.1002/ddr.v76.7
36. Cole SP. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J Biol Chem. 2014;289(45):30880–30888. doi:10.1074/jbc.R114.609248
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.