Back to Journals » Cancer Management and Research » Volume 12
MiR-548d-3p Promotes Gastric Cancer by Targeting RSK4
Authors Liang H, Hu C , Lin X , He Z, Lin Z, Dai J
Received 25 August 2020
Accepted for publication 4 December 2020
Published 24 December 2020 Volume 2020:12 Pages 13325—13337
DOI https://doi.org/10.2147/CMAR.S278691
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Bilikere Dwarakanath
Hui Liang,* Cong Hu,* Xu Lin, Zhuocheng He, Zhiwen Lin, Jun Dai
General Surgery Department, Zhuhai People’s Hospital (Zhuhai Hospital Affiliated with Jinan University), Zhuhai, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hui Liang
General Surgery Department, Zhuhai People’s Hospital (Zhuhai Hospital Affiliated with Jinan University), No. 79 Kangning Road, Xiangzhou District, Zhuhai 519000, Guangdong, People’s Republic of China
Tel +86 0756-2158341
Email [email protected]
Purpose: Previous studies have demonstrated that RSK4 inhibits the proliferation of gastric cancer cells and the occurrence of tumors. However, to date, studies involving microRNAs (miRNAs) that target RSK4 have rarely been reported. Thus, this study aimed to investigate the miRNAs that target RSK4.
Materials and Methods: We screened miRNAs related to RSK4 in miRDB, microT-CDS, TargetScan, and mirDIP databases and found 18 miRNAs. We chose miR-548d-3p for follow-up research, identified the interaction site in RSK4 by comparing the sequence, and mutated it. Thereafter, we used the dual-luciferase reporter system, real-time PCR (RT-PCR), and Western blotting to assess the effect of miR-548d-3p on RSK4. The proliferation, apoptosis, migration, and invasion of gastric cancer cells were evaluated using MTT assay, propidium iodide (PI), EdU, annexin V-FITC/PI apoptosis detection kit, wound healing assay, and transwell assay after overexpression of miR-548d-3p and RSK4. Finally, a nude mouse tumorigenesis experiment was conducted to explore the role of RSK4-targeting miR-548d-3p in tumorigenesis.
Results: miR-548d-3p negatively regulated the expression of RSK4, resulting in suppressed apoptosis, enhanced proliferation, migration, and invasion of gastric cancer cells, and accelerated tumor growth. In addition, an increase in miR-548d-3p expression enhanced the mRNA levels of CDK2, cyclin A1, cyclin D1, Bax, Bcl-2, N-cadherin, and Vimentin, and decreased E-cadherin mRNA levels by targeting RSK4.
Conclusion: miR-548d-3p promotes gastric cancer by lowering the expression of RSK4.
Keywords: microRNA, gastric cancer, RSK4, miR-548d-3p
Introduction
Gastric cancer is the fifth most common malignant tumor worldwide.1 Despite the decline in the incidence rate over the past century, its mortality rate in East Asia ranks third among cancer-related deaths.1–4 The main reason for the poor prognosis of gastric cancer is the lack of early diagnostic methods.3 The disease is usually diagnosed when tissue infiltration and distant metastasis have occurred, resulting in an average 5-year survival rate of less than 20% for patients in most parts of the world.1,3,5 Therefore, gastric cancer is still a clinical challenge for which effective early diagnosis targets and new therapeutic targets must be detected.1 Exploring the genes that regulate gastric cancer may assist in its early diagnosis and treatment.3,6,7
MicroRNA (miRNA) is a short single-stranded RNA molecule that can regulate the expression of genes and proteins.8–10 miRNA binds to the 3ʹ-untranslated region (UTR) of mRNA, increasing mRNA degradation and leading to downregulation of protein expression in the cytoplasm.8,11 Therefore, the expression of certain miRNAs is highly correlated with cancer.8,12 For example, miR-548d inhibits the progression of pancreatic cancer, miR-21 contributes to the occurrence of lung cancer, highly expressed miR-21 has been verified as important diagnostic markers for patients with breast and colon cancer, miR-200c and ovarian cancer are related, and the combination of hsa-miR-375 and hsa-miR-142-5p can be used to predict the risk of recurrence in patients with gastric cancer.8,13–17
RSK4 belongs to the p90 ribosomal S6 kinases (RSKs) family, and is known as RPS6KA6, which is involved in cell proliferation, invasion, and metastasis.1,18–23 RSK4 has a tumor-suppressing effect in colon, breast, and kidney cancer.1,24–27 Overexpressed RSK4 suppresses cell growth, invasion, and metastasis.1,28,29 Moreover, some researchers recommend RSK4 as a marker for evaluating the prognosis of patients.1,30 However, in some breast cancer tissues, the expression of RSK4 is higher than that in neighboring tissues, and whether RSK4 exerts a carcinogenic or a tumor suppressor effect depends on many factors.1,31,32 Moreover, our previous study has shown that the expression of RSK4 has an inhibitory effect on the progression of gastric cancer.1 However, reports on miRNAs that target RSK4 in gastric cancer are limited. The present study aimed to explore the biological functions of RSK4-targeting miRNAs in gastric cancer to provide a basis for its role in tumor therapy.
Materials and Methods
Cell Lines
Stomach cancer cell lines, HGC-27 (IMMOCELL, catalog number: IM-H085, Xiamen, China) and MGC-803 (IMMOCELL, catalog number: IM-H086, Xiamen, China), and 293T (IMMOCELL, catalog number: IM-H086, Xiamen, China) were cultured in Dulbecco’s modified eagle medium (DMEM, IMMOCELL, catalog number: IMC-201, Xiamen, China) supplemented with 10% fetal bovine serum (FBS, IMMOCELL, catalog number: IMC-101, Xiamen, China) and 1% penicillin-streptomycin (IMMOCELL, catalog number: IMC-601, Xiamen, China).
Screening of miRNAs That Target RSK4
The miRNA sequences that are related with RSK4 were downloaded from microT-CDS, miRDB, TargetScan, and mirDIP databases, and analyzed the distribution of these miRNAs in these databases using the UpSetR package in R software (version 3.5.1). Using survminer package (version 0.4.8) in R software, patients with gastric cancer from The Cancer Genome Atlas (TCGA) were divided into two groups: miR-548d-3p high expression and miR-548d-3p low expression. The Kaplan–Meier (KM) curve for the miR-548d-3p level and the disease-free survival was plotted using survival package (version 3.1–12) in R software.
Plasmid Construction
pCDH-EF1α-T2A-puro vector (Anti-HeLa BioTech, Xiamen, China) was used to prepare the plasmid expressing RSK4 (Gene ID: 27,330), named RSK4 plasmid, pCDH-EF1α-T2A-blasticidin vector (Anti-HeLa BioTech, Xiamen, China) and miR-548d precursor were used to prepare the plasmid expressing miR-548d, named miR-548d plasmid, and pmriGLO vector (Anti-HeLa BioTech, Xiamen, China) was used to construct the plasmid expressing wild-type or mutant 3ʹUTR of RSK4, named 3ʹUTR WT and 3ʹUTR MUT, respectively. The primers were designed by DNAMAN 10.0 and are listed in Table 1.
 |
Table 1 Primers for Plasmid Construction |
Dual-Luciferase Reporter Assay
HGC-27 and MGC-803 were used to performed dual-luciferase reporter assay through transfected 3ʹUTR WT or 3ʹUTR MUT with the negative control of miR-548d-3p mimic (mimic NC) or miR-548d-3p mimic for 48 h using Exfect 2000 Transfection Reagent (Vazyme, catalog number: T202-01, Nanjing, China) in accordance with the manufacturer’s instructions. The dual Luciferase Reporter Assay Kit was bought from Vazyme (catalog number: DL101-01, Nanjing, China).
Construction of Cell Lines Stably Overexpressing RSK4 or/and miR-548d
RSK4 plasmid, miR-548d plasmid, pCDH-EF1α-T2A-puro vector or pCDH-EF1α-T2A- blasticidin vector, and the suitable packaging plasmid (pMD2G, pspax2) were co-transfected into 293T cells using Lipofectamine 2000 (Invitrogen, catalog number: 11,668,019, Waltham, Massachusetts, USA) to prepare lentiviral particles. After 48 h, the lentivirus-containing medium was collected, and the particles were termed RSK4 lentivirus, miR-548d lentivirus, pCDH-EF1α-T2A-puro lentivirus, and pCDH-EF1α-T2A-blasticidin lentivirus, respectively.
HGC-27 cells were classified into Vector, miR-548d, and miR-548d+RSK4 cells. Vector cells were infected with pCDH-EF1α-T2A-blasticidin lentivirus, whereas miR-548d and miR-548d+RSK4 cells were infected with miR-548d lentivirus. After 48 h, the medium was replaced with fresh medium containing 15.0 μg/mL blasticidin S (Gibco, catalog number: A1113902, Shanghai, China). After 2 weeks, the cells were collected for analysis of miR-548d levels. When the levels of miR-548d in miR-548d and miR-548d+RSK4 cells were significantly higher than those in Vector cells, miR-548d and Vector cells were infected with pCDH-EF1α-T2A-puro lentivirus, and miR-548d+RSK4 cells were infected with RSK4 lentivirus. After 48 h, the medium was replaced with fresh medium containing 2.5 μg/mL puromycin (Gibco, catalog number: A1113803, Shanghai, China). After 1 week, cells were collected for miR-548d and RSK4 level analysis.
Real-Time PCR (RT-PCR)
RNA isolation Total RNA Extraction Reagent (Vazyme, catalog number: R401-01, Nanjing, China) was used to extract total RNA from cells or tissues. After reverse transcription of RNA into cDNA, RT-PCR was performed using cDNA as described previously.33 Primers for RT-PCR, designed using DNAMAN 10.0, are presented in Table 2.
 |
Table 2 Primers for RT-PCR |
Western Blotting
Ice-cold RIPA buffer (Cell Signaling Technology, Catalog No.: 9806, Shanghai, China) was used to extract proteins from cells or tissues. After protein quantification with BCA Protein Quantification Kit (Vazyme, Catalog No.: E112-01, Nanjing, China), Western blot analysis was performed as previously described.33 Anti-RSK4 antibody (Abcam, catalog number: ab76117, 1:1000, Shanghai, China) and anti-GAPDH antibody (Abcam, catalog number: ab8245, 1:2000, Shanghai, China) were used as primary antibodies. Goat anti-mouse IgG (Abcam, catalog number: ab6789, 1:2000, Shanghai, China) and goat anti-rabbit IgG (Abcam, catalog number: ab6721, 1:2000, Shanghai, China) were used as secondary antibodies conjugated with horseradish peroxidase.
Cell Proliferation Analysis
MTT, EdU, and cell cycle assays were used to analyze the cell proliferation. After HGC-27 and MGC-803 cell lines were transfected mimic NC or miR-548d-3p mimic with RSK4 plasmid or the negative control for 24 h (for EdU assay and cell cycle assay) or different time points: 24, 48, and 72 h (for MTT assay). MTT (Beyotime, catalog number: ST316, Shanghai, China), EdU (Beyotime, catalog number: ST067, Shanghai, China), and propidium iodide (PI) (Beyotime, catalog number: ST511, Shanghai, China) were used to detect cell proliferation as previously described.33
Apoptosis Assay
Cells treated as described above were used to perform apoptosis assay with annexin V-FITC/PI apoptosis detection kit purchased from Beyotime (catalog number: C1062M, Shanghai, China) as previously described.33
Cell Migration and Invasiveness Analysis
After cell treatment, wound healing assay, Matrigel-free transwell plates, transwell plates with Matrigel, and transwell assay were performed for cell migration and invasiveness analyses.33
Nude Mouse Tumorigenicity Assay
HGC-27 cells stably overexpressing miR-548d or both miR-548d and RSK4 were used for tumor formation experiments in nude mice. Fifteen nude mice purchased from Shanghai Experimental Animal Research Center (Shanghai, China) were divided equally into three groups: Vector, miR-548d, and miR-548d+RSK4. Nude mice in the Vector group were injected subcutaneously with negative control HGC-27 cells (Vector cells), nude mice in the miR-548d group were injected subcutaneously with HGC-27 cells stably and highly expressing miR-548d (miR-548d cells), and nude mice in the miR-548d+RSK4 group were injected subcutaneously with HGC-27 cells stably and highly expressing of miR-548d and RSK4 (miR-548d+RSK4 cells). The tumor size was measured on the 18th day after the nude mice were injected with the cells, thereafter, tumor size was measured every 4 days. On the 38th day after the injection of the cells, the nude mice were euthanized in a euthanasia chamber with carbon dioxide at a flow rate of 30–70% chamber volume per minute. Subsequently, the tumor was removed and weighed, and the levels of miR-548d and RSK4 were measured.
Statistical Analysis
Student’s t-test (unpaired) and analysis of variance (ANOVA) were used to perform comparisons between two groups and comparisons between multiple groups, respectively, using SPSS software (version 22.0). P < 0.05 indicates a significant difference. GraphPad Prism software (version 8.2.1) was used to draw the graph.
Results
MiR-548d-3p Negatively Regulates RSK4
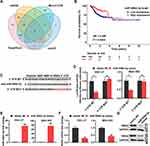
To screen for miRNAs that target RSK4, miRNAs related to RSK4 were downloaded from the microT-CDS, miRDB, TargetScan, and mirDIP databases. Eighteen miRNAs associated with RSK4 were present in all four databases, including miR-548d-3p (Figure 1A). Furthermore, KM curve analysis suggested that the level of miR-548d-3p is associated with disease-free survival of patients with gastric cancer (Figure 1B). Subsequently, we identified the site of RSK4 targeted miR-548d-3p and mutated it (Figure 1C). The site was verified by dual-luciferase reporter assay, and it was found that miR-548d-3p suppressed RSK4 transcription (Figure 1D). RT-PCR and Western blot analysis showed that miR-548d-3p also reduced RSK4 mRNA and protein levels (Figure 1E–G). These findings indicate that miR-548d-3p inhibits RSK4 expression.
MiR-548d-3p Promotes the Proliferation of Gastric Cancer Cells by Decreasing the Expression of RSK4
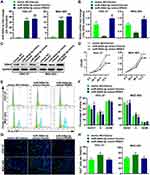
To investigate the effect of RSK4-targeting miR-548d-3p on the biological process of gastric cancer cells, we co-transfected miR-548d-3p mimic and RSK4 plasmids into HGC-27 and MGC-803. The results showed that miR-548d-3p attenuated the level of RSK4, and after supplementing RSK4, the level of RSK4 was restored (Figure 2A–C). MTT, EdU and cell cycle analysis revealed that overexpression of miR-548d-3p increased cell proliferation and accelerated mitosis, while reparative RSK4 suppressed the cell proliferation induced by miR-548d-3p (Figure 2D–H). These results indicate that miR-548d-3p enhances proliferation by targeting RSK4 in gastric cancer cells.
RSK4-Targeting miR-548d-3p Weakens Apoptosis and Strengthens Cell Migration and Invasion
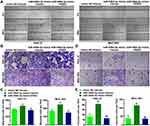
Next, we investigated the role of miR-548d-3p in apoptosis, migration, and invasion. By using the annexin V-FITC/PI apoptosis detection kit, wound healing assay, and transwell assay, it was found that overexpression of miR-548d-3p reduced the apoptosis rate and increased the number of migrating and invading cells, while supplementing the RSK4 heightened apoptosis rate and led to a decline in the number of migrating and invading cells (Figures 3 and 4). These data suggest that miR-548d-3p suppresses RSK4 expression, weakening apoptosis and intensifying the ability of cell migration and invasion.
RSK4-Targeting miR-548d-3p Regulates the mRNA Levels of Molecules Related to Cell Cycle, Apoptosis, and Epithelial-Mesenchymal Transition
We further verified the regulatory effects of miR-548d-3p on molecules related to cell cycle, apoptosis, and epithelial–mesenchymal transition. Overexpression of miR-548d-3p increased the mRNA levels of CDK2, cyclin A1, cyclin D1, Bax, Bcl-2, N-cadherin, and Vimentin, and decreased the mRNA level of E-cadherin. After supplementing RSK4, these effects of miR-548d-3p were alleviated (Figure 5), thus indicating that miR-548d-3p regulates the expression of other molecules by inhibiting the expression of RSK4, thereby regulating the cell cycle, cell apoptosis and cell epithelial–mesenchymal transition.
MiR-548d Promotes Tumor Growth by Targeting RSK4
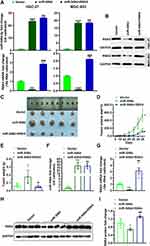
To investigate the RSK4-regulating role of miR-548d in tumors, we constructed cell lines with stable overexpression of miR-548d or stable overexpression of both miR-548d and RSK4 (Figure 6A and B). The results showed that the tumors formed in the miR-548d group were larger than those in the negative control and the miR-548d+RSK4 groups (Figure 6C) and had the fastest growth rate (Figure 6D). The weight of tumors formed in the miR-548d group was also higher than that in the other two groups (Figure 6E). The RT-PCR and Western blot experiments proved that in the formed tumors, the miR-548d levels of the miR-548d and the miR-548d+RSK4 groups were similar, while the RSK4 mRNA and protein levels of the miR-548d group were significantly lower than those of miR-548d+RSK4 group (Figure 6F–I). These results indicate that miR-548d negatively regulates RSK4 to accelerate tumor growth.
Discussion
Gastric cancer is a common malignant tumor and one of the main causes of cancer-related deaths.3 Non-coding RNA is dysregulated in the process of carcinogenesis and is considered useful in the diagnosis and treatment of cancer.3,34 miRNAs, a type of non-coding small RNA with 18 to 25 nucleotides, promote mRNA degradation or inhibit translation by binding to 3ʹUTR of the target mRNA, which causes changes in miRNA levels to seriously affect physiological and pathological processes.35–37 Furthermore, miRNA is involved in a variety of diseases, including cancer and cardiovascular diseases, and is considered an important therapeutic target for these diseases.35 miR-548 is involved in certain cancers and is considered a potential target for the treatment of cancers.38–40 The present study confirmed that miR-548d-3p targets RSK4 to increase the proliferation of gastric cancer cells and the occurrence of tumors in vivo, and may be a potential target for the treatment of gastric cancer.
Previous studies have revealed that miR-548d is an inhibitor of pancreatic cancer.17 miR-548d-3p inhibits osteosarcoma by down-regulating KRAS, and miR-548 inhibited the growth of breast cancer cells by down-regulating ECHS1.38,41 Interestingly, in breast cancer cells, miR-548d-3p down-regulates the expression of TP53BP2, thereby promoting the proliferation of breast cancer cells.42 These findings indicate that miR-548d-3p plays different roles in different situations. Nonetheless, our study found that miR-548d-3p promotes gastric cancer by inhibiting the expression of RSK4.
Overexpression of RSK4 in breast cancer cells reduces the proliferation, migration, and angiogenesis, and increases apoptosis.43 Compared with patients with hypermethylation of RSK4, patients with hypomethylation of RSK4 showed longer disease-free survival, indicating that hypomethylation of the RSK4 promoter may potentially contribute novel prognostic markers of breast cancer patients.43 In addition, overexpression of RSK4 reverses the resistance of human breast cancer cells to adriamycin through the PI3K/Akt signaling pathway.44 The expression of RSK4 is low in non-small cell lung cancer tissues and is related to the clinical and pathological staging of non-small cell lung cancer.45 RSK4, which is overexpressed in colorectal cancer cell lines, destroys the growth, metastasis, and invasion of tumor cells by inhibiting epithelial–mesenchymal transition. Similarly, in gastric cancer, reducing the expression of RSK4 inhibits cell apoptosis, leading to enhanced cell proliferation, migration, and invasion, and promotes tumor occurrence in vivo.1 These results indicate that RSK4 has a tumor suppressor effect in a variety of cancers. However, in lung adenocarcinoma, RSK4 is highly expressed, and is closely related to TNM stage, tumor diameter, lymph node metastasis, and distant metastasis, and is considered useful for the diagnosis of lung adenocarcinoma, as well as predicting the progression of this disease.46 Molecular studies showed that overexpression of RSK4 can enhance the invasion and migration ability of clear cell renal cell carcinoma cell lines by regulating the expression of CD44 and MMP-9.47 Thus, RSK4 plays different roles in different tumors, reflecting the diversity and complexity of RSK4 functions.
In a previous study, lowering RSK4 expression in breast cancer cells increased cyclin D1 mRNA levels and decreased E-cadherin mRNA levels.27 This is consistent with our results that overexpression of miR-548d-3p reduces RSK4 expression, thereby increasing cyclin D1 mRNA level and decreasing E-cadherin mRNA level. RSK4 overexpression has been reported to increase E-cadherin mRNA levels in SW480 and HCT116 cells.26 Similarly, we found that E-cadherin mRNA level was elevated after RSK4 supplementation.
Conclusion
We identified the RSK4-targeting miRNA, miR-548d-3p, and investigated its function in gastric cancer. Our findings reveal that miR-548d-3p accelerates the proliferation of gastric cancer cells and tumorigenesis by reducing the expression of RSK4.
Abbreviations
miRNAs, microRNAs; RT-PCR, real-time PCR; UTR, untranslated region; RSKs, ribosomal S6 kinases; DMEM, Dulbecco’s modified Eagle medium; FBS, fetal bovine serum; ANOVA, analysis of variance.
Ethics Committee Approval
This study was approved by the Institutional Ethics Committee of Zhuhai People’s Hospital (Zhuhai Hospital Affiliated with Jinan University). All of the experiments using animals were performed in accordance with a protocol approved by the Animal Care and Use Committee of Jinan University.
Disclosure
The authors have declared that no competing interest exists.
References
1. Hu C, Dai J, Lin X, Meng Y, Liang H. Effect of RSK4 on biological characteristics of gastric cancer. Cancer Manag Res. 2020;12:611–619. doi:10.2147/CMAR.S238132
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
3. Liang M, Huang G, Liu Z, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019;52(5):e12661. doi:10.1111/cpr.12661
4. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713. doi:10.1158/1055-9965.EPI-13-1057
5. Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42(2):211–217.
6. Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20(38):13767–13774. doi:10.3748/wjg.v20.i38.13767
7. Bertuccio P, Chatenoud L, Levi F, et al. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009;125(3):666–673. doi:10.1002/ijc.24290
8. Jiang F, Shen XB. miRNA and mRNA expression profiles in gastric cancer patients and the relationship with circRNA. Neoplasma. 2019;66(6):879–886. doi:10.4149/neo_2018_181211N952
9. Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer - an emerging concept. EBioMedicine. 2016;12:34–42. doi:10.1016/j.ebiom.2016.09.017
10. Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64(5):311–336.
11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
12. Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–1187. doi:10.1016/j.cell.2012.02.005
13. Saito M, Schetter AJ, Mollerup S, et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin Cancer Res. 2011;17(7):1875–1882. doi:10.1158/1078-0432.CCR-10-2961
14. Chan M, Liaw CS, Ji SM, et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19(16):4477–4487. doi:10.1158/1078-0432.CCR-12-3401
15. Marchini S, Cavalieri D, Fruscio R, et al. Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol. 2011;12(3):273–285. doi:10.1016/S1470-2045(11)70012-2
16. Zhang X, Yan Z, Zhang J, et al. Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol. 2011;22(10):2257–2266. doi:10.1093/annonc/mdq758
17. Heyn H, Schreek S, Buurman R, Focken T, Schlegelberger B, Beger C. MicroRNA miR-548d is a superior regulator in pancreatic cancer. Pancreas. 2012;41(2):218–221. doi:10.1097/MPA.0b013e318224b701
18. Frödin M, Gammeltoft S. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol. 1999;151(1–2):65–77. doi:10.1016/S0303-7207(99)00061-1
19. Lara R, Seckl MJ, Pardo OE. The p90 RSK family members: common functions and isoform specificity. Cancer Res. 2013;73(17):5301–5308. doi:10.1158/0008-5472.CAN-12-4448
20. Fogarty S, Hardie DG. C-terminal phosphorylation of LKB1 is not required for regulation of AMP-activated protein kinase, BRSK1, BRSK2, or cell cycle arrest. J Biol Chem. 2009;284(1):77–84. doi:10.1074/jbc.M806152200
21. Kang S, Elf S, Lythgoe K, et al. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest. 2010;120(4):1165–1177. doi:10.1172/JCI40582
22. Lara R, Mauri FA, Taylor H, et al. An siRNA screen identifies RSK1 as a key modulator of lung cancer metastasis. Oncogene. 2011;30(32):3513–3521. doi:10.1038/onc.2011.61
23. Buck M, Poli V, Hunter T, Chojkier M. C/EBPbeta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol Cell. 2001;8(4):807–816. doi:10.1016/S1097-2765(01)00374-4
24. Li Q, Jiang Y, Wei W, Ji Y, Gao H, Liu J. Frequent epigenetic inactivation of RSK4 by promoter methylation in cancerous and non-cancerous tissues of breast cancer. Med Oncol. 2014;31(1):793. doi:10.1007/s12032-013-0793-3
25. López-Vicente L, Armengol G, Pons B, et al. Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clin Cancer Res. 2009;15(14):4546–4553. doi:10.1158/1078-0432.CCR-08-3159
26. Ye Q, Wang X, Jin M, et al. Effect of RSK4 on biological characteristics of colorectal cancer. World J Surg Oncol. 2018;16(1):240. doi:10.1186/s12957-018-1474-7
27. Zhu J, Li QY, Liu JL, Wei W, Yang HW, Tang W. RSK4 knockdown promotes proliferation, migration and metastasis of human breast adenocarcinoma cells. Oncol Rep. 2015;34(6):3156–3162. doi:10.3892/or.2015.4291
28. Dümmler BA, Hauge C, Silber J, et al. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J Biol Chem. 2005;280(14):13304–13314.
29. Thakur A, Sun Y, Bollig A, et al. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin Cancer Res. 2008;14(14):4427–4436. doi:10.1158/1078-0432.CCR-08-0458
30. Cai J, Ma H, Huang F, et al. Low expression of RSK4 predicts poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2014;7(8):4959–4970.
31. Thakur A, Rahman KW, Wu J, et al. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol Cancer Res. 2007;5(2):171–181. doi:10.1158/1541-7786.MCR-06-0071
32. Sun Y, Cao S, Yang M, et al. Basic anatomy and tumor biology of the RPS6KA6 gene that encodes the p90 ribosomal S6 kinase-4. Oncogene. 2013;32(14):1794–1810.
33. Li D, Cheng X, Zheng W, Chen J. Glucosamine-6-phosphate isomerase 1 promotes tumor progression and indicates poor prognosis in hepatocellular carcinoma. Cancer Manag Res. 2020;12:4923–4935. doi:10.2147/CMAR.S250094
34. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379. doi:10.1146/annurev-biochem-060308-103103
35. Son GH, Kim Y, Lee JJ, et al. MicroRNA-548 regulates high mobility group box 1 expression in patients with preterm birth and chorioamnionitis. Sci Rep. 2019;9(1):19746. doi:10.1038/s41598-019-56327-9
36. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi:10.1038/nrg2290
37. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
38. Chen J, Yan C, Yu H, Zhen S, Yuan Q. miR-548d-3p inhibits osteosarcoma by downregulating KRAS. Aging. 2019;11(14):5058–5069. doi:10.18632/aging.102097
39. Ni XF, Zhao LH, Li G, et al. MicroRNA-548-3p and MicroRNA-576-5p enhance the migration and invasion of esophageal squamous cell carcinoma cells via NRIP1 down-regulation. Neoplasma. 2018;65(6):881–887. doi:10.4149/neo_2018_171206N803
40. Saffari M, Ghaderian SMH, Omrani MD, Afsharpad M, Shankaie K, Samadaian N. The association of miR-let 7b and miR-548 with PTEN in prostate cancer. Urol J. 2019;16(3):267–273.
41. Shi Y, Qiu M, Wu Y, Hai L. MiR-548-3p functions as an anti-oncogenic regulator in breast cancer. Biomed Pharmacother. 2015;75:111–116.
42. Song Q, Song J, Wang Q, et al. miR-548d-3p/TP53BP2 axis regulates the proliferation and apoptosis of breast cancer cells. Cancer Med. 2016;5(2):315–324. doi:10.1002/cam4.567
43. Huo H, Ye X, Yang H, Li Q, Jiang Y. RSK4 inhibits breast cancer cell proliferation and invasion in vitro, and is correlated with estrogen receptor upregulation in breast cancer. Oncol Rep. 2019;42(6):2777–2787.
44. Mei Y, Liao X, Zhu L, Yang H. Overexpression of RSK4 reverses doxorubicin resistance in human breast cancer cells via PI3K/AKT signalling pathway. J Biochem. 2020;167(6):603–611. doi:10.1093/jb/mvaa009
45. Li A, Liu D, Liu Y, et al. Study of RSK4 expression in patients with human non-small cell lung carcinoma. Ann Clin Lab Sci. 2018;48(4):484–489.
46. He Q, He R, Luo W, et al. Expression of RSK4 in lung adenocarcinoma tissue and its clinicopathological value: a study based on RNA-seq data and immunohistochemistry. Int J Clin Exp Pathol. 2017;10(12):11405–11414.
47. Ma J, Li M, Chai J, et al. Expression of RSK4, CD44 and MMP-9 is upregulated and positively correlated in metastatic ccRCC. Diagn Pathol. 2020;15(1):28. doi:10.1186/s13000-020-00948-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.