Back to Journals » OncoTargets and Therapy » Volume 7
miR-544a promotes the invasion of lung cancer cells by targeting cadherina 1 in vitro
Authors Mo X, Zhang F, Liang H, Liu M, Li H, Xia H
Received 1 February 2014
Accepted for publication 17 March 2014
Published 4 June 2014 Volume 2014:7 Pages 895—900
DOI https://doi.org/10.2147/OTT.S61695
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Xiaomei Mo,1,2,* Fenghua Zhang,2,* Hui Liang,2 Ming Liu,1 Huahui Li,3 Haiping Xia3
1Key Laboratory of Marine Drugs, Ministry of Education, School of Medicine and Pharmacy, Ocean University of China, 2Qingdao Women and Children Hospital, 3Department of Laboratory Medicine, Qingdao Municipal Hospital, Qingdao, People's Republic of China
*These authors contributed equally to this work
Objective: To find out the effect of miR-544a on the invasion of lung cancer cells and to explore the underlying molecular mechanisms.
Methods: Micro-ribonucleic acid (miRNA) expression in two different invasive lung cancer cell lines 95C (low invasive ability) and 95D (high invasive ability) was analyzed by miRNA microarray and real-time quantitative polymerase chain reaction (PCR); miR-544a mimic was transfected to 95C, and its invasion ability was detected by transwell migration assay; we predicted the candidate miRNA target genes by TargetScan (Whitehead Institute for Biomedical Research, Cambridge, MA, USA) software and verified the target genes by Western blot.
Results: The expression of miR-544a was significantly increased in 95D in miRNA microarray and quantitative PCR tests (P<0.05). After being transfected with miR-544a mimic, the invasion ability of 95C was enhanced (P<0.01). Moreover, transfection with miR-544a inhibitor decreased the invasion ability of 95D (P<0.01). miR-544a possibly combined with CDH1 (E-cadherin) predicted by the TargetScan analysis. 95C with miR-544a mimic reduced the expression of CDH1 and improved the expression of vimentin, while 95D with miR-544a inhibitor improved the expression of CDH1 and reduced the expression of vimentin.
Conclusion: miR-544a can promote the invasion of non-small cell lung cancer by downregulation of CDH1 and upregulation of vimentin.
Keywords: NSCLC, non-small cell lung cancer, E-cadherin, microRNA, EMT
Introduction
Non-small cell lung cancer (NSCLC) occupies 80% of all types of lung cancer,1 and many cancer patients die of cancer invasion or metastasis. Therefore, it is necessary to do further study on the molecule markers that play an important role in the invasion or metastasis in NSCLC.
Mature micro-ribonucleic acids (miRNAs) consist of 22 nucleotides, and as negative regulators of gene expression, mainly recognize the complementary sequences in the 3′ untranslated regions (UTRs) of their target messenger RNAs.2 Many studies have revealed that miRNA not only participates in biological processes such as cell cycle, aging, and death but also is a hallmark of several pathological conditions, including cancer, exerting a causal role, as oncogenes or tumor suppressors. For example, miR-544a levels are reduced in glioma3 and invasive ductal carcinoma.4 miR-544a can be seen as the molecular marker of glioma and tumor suppressor. miR-544a exerts its carcinogenic factor, promoting cell cycle and cell proliferation of stomach cancer, by reducing IRX1, one tumor suppressor gene.5 However, there has been little study on the role of miR-544a in NSCLC metastasis and invasion. Invasion and metastasis usually occur even after complete lung cancer resection; therefore, two different invasive lung cancer cell lines (95C and 95D) were used to explore the miR-544a levels, and predict the target gene and its potential downstream proteins. This study will provide insights into the role of miR-544a in lung cancer invasion and metastasis and the possible molecular mechanisms.
Materials and methods
Cell lines and reagents
The 95D and 95C cells were purchased from the American Type Culture Collection and cultured in Dulbecco’s Modified Eagle’s Medium with 10% fetal bovine serum (FBS). The 95D cells showed higher invasion, while the 95C cells showed lower invasion. Moloney murine leukemia virus reverse transcriptase (MMLV-RT) was purchased from Toyobo Corporation (Osaka, Japan).
miRNA array
106–107 95D or 95C cells in logarithmic phase, conventional cell culture, and total RNA were extracted with TRIzol® (Life Technologies, Carlsbad, CA, USA). miRNA array was performed in Beijing Bo Ao Capitalbio Corporation with 40 μg total RNA. Bo Ao miRNA array included 469 probes. U6 and transfer RNA were used as inner control, and eight synthetic RNAs (20–30 nucleotides) were used as exogenous control. Hex was used as positive control, and 50% DMSO (dimethyl sulfoxide) as negative control (NC). Loess normalization was used to normalize the miRNA array data. Every RNA sample was performed in duplicate.
Bioinformatics analysis
The target gene was predicted by TargetScan (Whitehead Institute for Biomedical Research, Cambridge, MA, USA) software.
Luciferase assays
LightSwitch™ (Promega, Fitchburg, WI, USA) luciferase assay reagents are from Promega Corporation (Fitchburg, WI, USA). miRNA NC and miR-544a mimic combine with E-cadherin (CDH1) 3′UTR or CDH1 mutated 3′ UTR and are transfected to 293T cells for 24 hours, respectively. Assay reagents were added to each well according to the manufacturer’s instructions (Promega Corporation). Firefly luciferase (FLuc) and renilla luciferase (RLuc) were counted by a chemiluminescence meter (Promega Corporation). Luciferase expression was recorded as relative light units (RLuc/FLuc) to estimate whether CDH1 is the target gene of miR-544a.
Quantitative polymerase chain reaction (PCR)
Total RNA was extracted from 95C or 95D with TRIzol and reverted to complementary deoxyribonucleic acid (cDNA) by MMLV-RT. Quantitative PCR was performed using a PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) with the use of SYBR® Green I Premix Ex Taq™ (Takara Bio Inc, Shiga, Japan). Specific primers for miR-544a were designed by Rui Bo Company (Guangzhou, People’s Republic of China). Reagents include 2× Mix SYBR Green I 10 μL, 10 pM primers 0.25 μL, cDNA 1 μL, and double-distilled H2O up to 20 μL. The reaction protocol included an initial step of 120 seconds at 95°C. Each PCR cycle involved denaturation (95°C, 30 seconds), annealing (60°C, 35 seconds), and extension (72°C, 20 seconds) for 40 cycles. Fluorescence was measured at each cycle. The relative fold change of miR-544a expression was quantified as 2−ΔΔCt, where ΔΔCt was Ct (target genes) – Ct (housekeeping genes). We selected small nuclear RNA U6 as housekeeping gene. The nucleotide sequence of the upstream primer is TGGCACCCAGCACAATGAA; and the downstream primer, CTAAGTCATAGTCCGCCTAGAAGCA.
Transfection
95C was transfected with miR-544a mimic and NC, respectively, while 95D was transfected with miR-544a inhibitor and NC, according to the manufacturer instructions for Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA): 1×105 95C or 95D, 50 μL 1× buffer, 2.5 μL miR-544a mimic, miR-544a inhibitor, or NC. After being transfected for 36 hours, the cells were applied to transwell migration assay and Western blot.
Transwell migration assay
Put Matrigel™ (BD Biosciences, San Jose, CA, USA) at 4°C overnight. Dilute Matrigel with serum-free Roswell Park Memorial Institute (RPMI)-1640 at a 1:9 dilution. Add 50μL diluted Matrigel to each transwell hole and put it at 37°C for 3~4 hours.A total of 5×104 transfected cells (95C transfected with miR-544a mimic and NC; 95D with miR-544a inhibitor and NC) suspended in 200 μL serum-free RPMI-1640 medium was placed into the upper chamber. Outside the upper chamber, 500 μL 10% FBS-RPMI-1640 medium was added. After 24 hours of incubation, cells were washed with PBS three times, and cells remaining on the upper membrane were carefully removed. Cells that had migrated through the membrane were fixed with methanol and stained with hematein for 5 minutes. Finally, the migrated cells were imaged and counted at 10× magnification using a Leica DC 300F microscope (Olympus Corporation, Tokyo, Japan).
Western blot
95C-NC, 95C-miR-544a-mimic, 95D-miR-544a-inhibitor, and 95D-NC were lysed by 100 μL radioimmunoprecipitation assay buffer (RIPA; Sigma-Aldrich, St Louis, MO, USA). The protein concentration was detected by bicinchoninic acid (BCA) protein assay kit (HyClone-Pierce, Logan, UT, USA). Protein (20 μg) was loaded on 12% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) (120 V, 1.5 hours), transferred to PVDF (polyvinylidene difluoride) membrane and blotted by 5% skim milk powder for 2 hours. Membranes were probed with primary antibodies (1:10,000) of CDH1 (Abcam, Cambridge, MA, USA), vimentin (Santa Cruz Biotechnology, Inc., Dallas, TX, USA), α-tubulin (Santa Cruz Biotechnology, Inc.), and horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology, Inc.). Protein expression was quantitatively assessed using a HRP-ECL scanner (Lenovo, Beijing, People’s Republic of China).
Statistical analysis
Student-Newman-Keuls-q (SNK-q) is used to compare pairs of means of multiple data, and Student’s t-test is used for the difference in only two means. One-way ANOVA (analysis of variance) with SNK-q test for multiple comparisons was used to analyze the data from the transwell migration assay and Western blot using SPSS 15.0 (SPSS Inc., Chicago, IL, USA) software. Student’s t-test was used to analyze the results of quantitative PCR. Data are shown as mean ± standard deviation. Differences of P<0.05 were considered statistically significant.
Results
95C and 95D expressed different levels of miR-544a
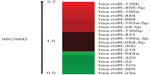
In the miRNA array, we analyzed the miR-544a levels in 95C and 95D cells, and the results showed that the ratio of miR-544a levels in 95C and 95D was very low, indicating that the level of miR-544a in 95D was higher than that in 95C (Figure 1). To verify the results of the miRNA array, quantitative PCR was carried out, and the results showed that the relative level of miR-544a in 95D was almost threefold that in 95C cells (Figure 2 and Table 1). All of these results showed that there is a higher level of miR-544a in 95D than in 95C cells.
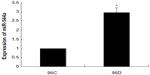  | Figure 2 miR-544a levels in 95C and 95D by quantitative polymerase chain reaction. The relative level of miR-544a in 95D was almost three times that in 95C cells. |
CDH1 is one of the target genes of miR-544a
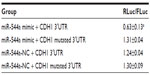
TargetScan was used to predict the target gene of miR-544a, and the results showed that miR-544a possibly combined with CDH1 (Figure 3). CDH1 is one of the cell adhesion molecules on cell membranes, maintaining cell integrity and exerting the ability of cell contact inhibition. To verify the interaction between CDH1 and miR544a, luciferase assay was carried out. As shown in Figure 4 and Table 2, miR-544a mimic could combine with CDH1 3′UTR and inhibit the expression of the reporter gene (0.63±0.13). When CDH1 3′UTR mutated, miR-544a mimic could not combine with CDH1 mutated 3′UTR and the expression of the reporter gene (1.31±0.04) increased (q=10.12, P<0.01). These results suggest that CDH1 is one of the target genes of miR-544a.
miR-544a enhances the migration of NSCLC cells
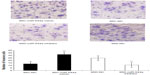
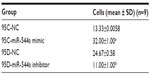
To study the effect of miR-544a on cancer cell migration, a transwell migration assay was performed, and the results are shown in Figure 5. After transfection with miR-544a mimic, migrated cells numbered more than that of 95C-NC (q=18.67, P<0.01). When cells were treated with 95D-miR-544a inhibitor, migrated cells numbered less than that of 95D-NC (q=18.67, P<0.01) (Table 3). These results suggest that miR-544a could enhance the migration of NSCLC cells.
miR-544a downregulates CDH1
To investigate whether miR-544a could affect the expression of CDH1, Western blot was performed. In cells treated with 95C-miR-544a mimic, the level of CDH1 decreased and that of vimentin increased compared with 95C-NC (P<0.01). When treated with 95D-miR-544a inhibitor, the level of CDH1 increased while that of vimentin reduced significantly (q-value was 18.67 and 5.99, respectively; P<0.01; Figure 6). The results suggested that miR-544a could downregulate CDH1 and upregulate vimentin in 95C cells.
Discussion
miRNA participates in tumor invasion and metastasis,6 but its mechanism is not very clear. In this study, we focused on two cell lines of NSCLC (95C and 95D), whose invasive ability is completely different. In the miRNA array assay, the miR-544a level was higher in 95D cells, which possess higher invasive ability. To avoid false positive results of the miRNA array, quantitative PCR was carried out to verify these results. Quantitative PCR also confirmed that the relative level of miR-544a in 95D was higher. In the transwell migration assay, the migration ability of 95C transfected with miR-544a mimic was higher, but that of 95D transfected with miR-544a inhibitor was lower. TargetScan is one of the most common software products used to predict an miRNA target gene. TargetScan showed that CDH1 may be the target gene of miR-544a. Luciferase assay also revealed that miR-544a could combine with CDH1 3′UTR and inhibit the expression of the reporter gene. In our previous work, we found that glycogen synthase kinase 3β is another target of miR-544a and is regulated by 95D cells.7
It is accepted that downregulation of CDH1 and upregulation of vimentin can promote the invasion of lung cancer. In the present study, we found that in 95C transfected with miR-544a mimic, CDH1 levels reduced and vimentin levels increased. But in 95D transfected with miR-544a inhibitor, CDH1 levels increased and vimentin levels reduced. Downregulation of CDH1 is considered as the mark of epithelial–mesenchymal transition (EMT), which plays an important role in the process of tumor metastasis.8 EMT makes tumor’s acquire invasive and metastatic ability. We found that in the process of EMT, CDH1 levels reduced and vimentin levels increased, which is consistent with the findings of Thiery and Lim.9 However, further in vivo studies are needed to confirm the role of miR-544a in invasion and metastasis of NSCLC and its regulation on CDH1 and vimentin.
In conclusion, our current study provided novel evidence that higher expression of miR-544a significantly suppresses CDH1, resulting in the promoting of the invasion and metastasis of NSCLC in vitro. The regulation of miR-544a on CDH1 provides novel insight into the metastasis of NSCLC, especially with respect to invasion and metastasis in vitro, and also represents a new potential therapeutic target for the treatment of NSCLC.
Disclosure
The authors report no conflicts of interest in this work.
References
Lam WK, Watkins DN. Lung cancer: future directions. Respirology. 2007;12(4):471–477. | |
Vimalraj S, Miranda PJ, Ramyakrishna B, et al. Regulation of breast cancer and bone metastasis by microRNAs. Dis Markers. 2013;35(5): 369–387. | |
Ma R, Zhang G, Wang H, et al. Downregulation of miR-544 in tissue, but not in serum, is a novel biomarker of malignant transformation in glioma. Oncol Lett. 2012;4(6):1321–1324. | |
Haga CL, Phinney DG. MicroRNAs in the imprinted DLK1-DIO3 region repress the epithelial-to-mesenchymal transition by targeting the TWIST1 protein signaling network. J Biol Chem. 2012;287(51): 42695–42707. | |
Zhi Q, Guo X, Guo L, et al. Oncogenic miR-544 is an important molecular target in gastric cancer. Anticancer Agents Med Chem. 2013;13(2):270–275. | |
Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15(6): 546–554. | |
Mo XM, Li HH, Li YT. Down regulation of GSK3β by miR-544a to maintain self-renewal ability of lung cancer stem cells. Oncol Lett. In press 2014. | |
Chen Y, Sun Y, Chen L, et al. miRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 2013;7(5): 1579–1584. | |
Thiery JP, Lim CT. Tumor dissemination: an EMT affair. Cancer Cell. 2013;23(3):272–273. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.