Back to Journals » Cancer Management and Research » Volume 11
miR-526b-3p serves as a prognostic factor and regulates the proliferation, invasion, and migration of glioma through targeting WEE1
Authors Wu M, Li X, Liu Q, Xie Y, Yuan J, Wanggou S
Received 26 October 2018
Accepted for publication 17 January 2019
Published 11 April 2019 Volume 2019:11 Pages 3099—3110
DOI https://doi.org/10.2147/CMAR.S192361
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Ming Wu, Xuejun Li, Qing Liu, Yuanyang Xie, Jian Yuan, Siyi Wanggou
Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China
Background: MicroRNAs play important roles in cancer progression including glioma. In this study, we aimed to explore the expression pattern, prognostic potential, and functional role of miR-526b-3p in human glioma.
Materials and methods: The expression of miR-526b-3p in glioma tissues and the adjacent non-tumor tissues was determined by quantitative RT-PCR. The chi-square test was performed to evaluate the statistical associations between miR-526b-3p level and patient characteristics. The prognostic value of miR-526b-3p was analyzed by Kaplan–Meier and Cox regression analyses. The function of miR-526b-3p was analyzed by MTT, colony formation assay, transwell assay, and flow cytometry analysis in vitro. The binding between miR-526b-3p and predicted target WEE1 was verified using dual luciferase assay and Western blot analysis.
Results: We found that miR-526b-3p expression was significantly downregulated in both glioma tissues and cell lines. Downregulation of miR-526b-3p was significantly associated with advanced WHO grade, lower KPS score, and inferior patient outcomes. Functional investigation indicated that overexpression of miR-526b-3p suppressed cell proliferation, migration, and invasion, and promoted apoptosis in glioma cell lines. Mechanically, WEE1 was identified as direct targets of miR-526b-3p and overexpression of WEE1 significantly suppressed the levels of WEE1. Moreover, re-introduction of WEE1 abrogates the suppression of motility and invasiveness induced by miR-526b-3p in glioma cells.
Conclusion: These findings indicate that miR-526b-3p may target WEE1 and inhibit glioma tumorigenesis and progression.
Keywords: miR-526b-3p, glioma, tumorigenesis, prognosis, WEE1
Introduction
Glioma is a type of tumor that occurs in the brain and spinal cord and remains the most common brain tumor in adults.1,2 Glioma displays many malignant characteristics such as high invasiveness and resistance.3 Although surveillance and clinical treatment strategies have been improved, the median survival of patients in grade IV is still less than 1 year.4 This high lethality could be due to that surgery cannot remove the entire tumor without harming the healthy brain.5 In addition, symptoms of gliomas depend on which part of the central nervous system is affected, and glioma patients were frequently diagnosed at an advanced stage.6 In order to improve the prognosis of glioma patients, understanding the molecular signaling pathways and identification of novel biomarkers are very important.
MicroRNAs (miRNAs), highly conserved non-coding RNAs, modulate the expression of mRNAs by binding to their 3ʹ-untranslated region (3ʹ-UTR).7 Recent evidences show that miRNAs play crucial roles in various biological processes such as differentiation, proliferation, invasion, and apoptosis, and their aberrant expression is thought to be relevant to the progression of various diseases, including cancers.8–10 With the deepening of research, some miRNAs are tumor suppressors while others serve as oncogenes in cancer development, which suggests that they are considered as potential therapeutic targets.11,12 Due to the wide use of next-generation sequencing technology, numerous miRNAs have been identified.13,14 However, the function and potential mechanism of most of them remains largely unclear.
miR-526b-3p, a functional miRNA and located in 19q13.42, has been reported to be abnormally expressed in various tumors and plays an important role in the progression of these diseases.15,16 Generally speaking, miR-526b-3p was found to exhibit tumor suppressive role in several tumors, which revealed its potential role as a therapeutic target. To our best knowledge, the roles of miR-526b-3p in glioma have not been investigated. In this study, we first explored its expression, clinical significance, and function, and provided evidences that miR-526b-3p is a potential prognostic marker in glioma and functions as a tumor suppressor by targeting WEE1 which acted as an oncogene in glioma.
Materials and methods
Patients and tissue specimens
Human glioma tissue samples and corresponding adjacent normal brain tissues were obtained from 158 patients with glioblastoma at Xiangya Hospital, Central South University from March 2010 to April 2013. All the tissues collected from the patients were immediately frozen in liquid nitrogen and stored at −80°C after surgery. The patients did not receive any radiotherapy or chemotherapy before surgery. The histological grade of all glioma tissue samples was classified by experienced pathologists using WHO criteria. Written informed consent was received from the participants. The research protocols were approved by the Ethics Committee of Xiangya Hospital, Central South University. The study was conducted in accordance with the Declaration of Helsinki. The patients’ clinical information is listed in Table 2.
 | Table 1 The primers for qRT-PCR |
 | Table 2 Association between miR-526b-3p expression and clinicopathological features of 158 human gliomas |
Cell lines and cell transfection
Human glioma cell lines (T98G, U251, LN18, LN229, and A172) were obtained from the Chinese Academy of Sciences Cell Bank (Xuhui, Shanghai, China), and normal human astrocytes (NHAs) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cells were all cultured in RPMI-1640 medium (BasalMedia Technologies, Pudong, Shanghai, China) containing 10% of fetal bovine serum (FBS; HyClone, Logan, Utah, USA) and antibiotics (100 U/mL streptomycin and 100 U/mL penicillin) at 37°C with 5% CO2.
The negative control (NC) mimics and miR-526b-3p mimics were all purchased from RiboBio Co., Ltd. (Guangzhou, Guangdong, China). The pcDNA3.1-WEE1 plasmid which led to express WEE1 was constructed by Generay Biotechnology Co., Ltd. (Songjiang, Shanghai, China). Cell transfection was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s protocols.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured cells or clinical samples using TRIeasy Total RNA Extraction Reagent (Yeasen Biotechnology, Pudong, Shanghai, China) according to the manufacturer’s protocols. Afterward, the RNA was reversely transcripted to cDNA in a first-strand cDNA synthesis reaction with PrimeScript RT-PCR kit (Takara, Dalian, Liaoning, China). Then, qRT-PCR assays were conducted using All in One qPCR Mix kit (Applied GeneCopoeia Inc., Rockville, MD, USA) on an ABI 7500HT System (Applied Biosystems, Foster City, CA, USA). GAPDH was employed as an internal control for mRNA detection. For miRNA determination, miRNA Purification kit (Qiagen, Hilden, Germany) and TransScript Green miRNA Two-Step qRT-PCR SuperMix kit (TRANSGEN, Haidian, Beijing, China) were utilized. MiR-526b-3p was normalized to snRNA U6. Expression fold changes were calculated using 2−ΔΔCt method. All primer sequences are listed in Table 1.
Western blot analysis
Cells were collected and lysed using RIPA buffer (Meilun Biotechnology, Dalian, Liaoning, China) supplemented with protease inhibitor cocktail (Sigma Aldrich, St. Louis, MO, USA). Total protein concentrations of the cell lysate were assessed by 1× Bradford Assay kit (Bio-Rad Laboratories, Hercules, CA, USA). Subsequently, 20 μg protein samples were separated on a 10% SDS-PAGE by the use of a BioRad Bis-Tris Gel system (Bio-Rad, CA, USA), transferred onto polyvinylidene difluoride membranes (Millipore, Danvers, MA, USA), blocked with 5% non-fat milk for 1 hr at room temperature, and immunoblotted with primary antibodies. The primary antibodies against vimentin and N-cadherin were purchased from Abcam Co., Ltd. (Cambridge, MA, USA). The primary antibodies against GAPDH and WEE1 were purchased from Santa Cruz Biotechnology Co., Ltd. (Santa Cruz, CA, USA) and ProteinTech Co., Ltd. (Wuhan, Hubei, China). Then, the corresponding secondary antibodies were utilized to incubate with the membranes. The protein was then visualized using an ECL detection kit (Pierce, Rockford, IL, USA) as well as a Gel Imaging System (GE Healthcare, Madison, WI, USA).
Cell viability and colony formation assays
Cell proliferative abilities were tested by the Cell Counting Kit-8 (CCK-8; KeyGen Biotechnology, Nanjing, Jiangsu, China). In short, the pre-transfected U251 or LN299 cells were seeded into 96-well plates (Nunc, Roskilde, Denmark) with the amount of 3×103 cells per well. Then, 10 μL CCK8 reagent was added to each well at indicated time points. After maintaining for 1–2 hrs, a microplate reader (BioTek Instruments, Bio-Tek, Winooski, VT) was applied to evaluate the absorbance at a wavelength of 450 nm.
For colony formation assays, U251 or LN299 cells in the logarithmic growth phase were seeded in 6-well plates (Nunc, Roskilde, Denmark) at 37°C with 5% CO2 at a density of 400 cells per well. The cells were continued to be maintained at 37°C with 5% CO2 for approximately 2 weeks. At the end of monitored time, the cell colonies were fixed with ethanol, treated with 0.3% crystal violet solution (Sangong, Songjiang, Shanghai, China) for 30 mins, washed twice by deionized water, and photographed by an inverted microscope (IX71, Olympus, Tokyo, Japan).
Apoptosis assay
The Annexin V-FITC Apoptosis Detection kit purchased from DOJINDO Laboratories Co., Ltd. (Xuhui, Shanghai, China) was employed to assess the apoptosis of U251 and LN299 cells. In brief, the cells were collected, re-suspended in the binding buffer (400 μl), and subsequently incubated with 5 μL Annexin V-FITC as well as 5 μL propidium iodide (PI) after 24-hr transfection. Afterward, the cells were washed twice and the results were examined by an FACS Calibur system FACS AriaIII system (BD Biosciences, San Jose, CA, USA) and analyzed with FlowJo software (Version 7.6.1; Tree Star Inc., Ashland, Oregon, USA).
Transwell assays
Transwell assays were applied to evaluate the migratory and invasion capabilities of U251 and LN299 cells. The transwell migration assays were carried out using an 8-μm pore polycarbonate membrane chamber insert in a 24-well plate (BD Biosciences, Franklin Lakes, NJ, USA). For the invasion assays, the chamber inserts were pre-coated with 50 μL of Matrigel (200 mg/mL; BD Biosciences, Franklin Lakes, NJ, USA). Subsequently, cells were re-suspended in serum-free medium, plated into the upper chamber of inserts, and the medium containing 20% FBS was added into the lower chamber. After 24 hrs, the non-invading cells were gently removed with a cotton swab, and migratory or invasive cells attached to the lower surfaces of the chamber membranes were fixed in 100% methanol and stained with 0.3% crystal violet solution (Sangong, Songjiang, Shanghai, China). Then, the cells were counted and photographed by an inverted microscope (IX71, Olympus, Tokyo, Japan).
Dual luciferase reporter assays
Dual-Glo Luciferase Assay System kit (Promega, Madison, WI, USA) was utilized to examine the luciferase activity of U251 and LN299 cells. The wild-type 3ʹ‐untranslated region (UTR) or mutant 3ʹ-UTR sequence of WEE1 (WEE1 WT or WEE1 MUT) was sub-cloned into pGL3 Basic vector (Promega, Madison, WI, USA) by Vigene Biotechnology Co., Ltd. (Jinan, Shandong, China). The cells were seeded in 24-well plates and co-transfected with WEE1 WT or WEE1 MUT reporter vector together with NC mimic or miR-526b-3p mimic using Lipofectamine 2000 reagent (Invitrogen Co., Carlsbad, CA, USA) according to the manufacturer’s instructions. Following cultivation for 48 hrs, the transfected cells were collected and used for luciferase activity determination according to the protocols in the Dual-Glo Luciferase Assay System kit.
Data analysis
Differences in the results of two groups were evaluated using unpaired two-tailed Student’s t-test. The multi-group comparison was performed using one-way analysis of variance. The paired comparison was performed by SNK approach. Survival rates were calculated using the Kaplan–Meier method, and differences in survival curves were analyzed by log-rank tests. Multivariate Cox proportional hazards regression models were used to identify independent prognostic factors. Statistical analyses were performed with the SPSS 20.0 statistics software (SPSS, Inc., Chicago, IL, USA). A two-sided P-value<0.05 was considered statistically significant.
Results
MiR-526b-3p was downregulated in glioma tissues and cell lines
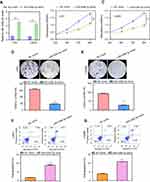
In order to investigate the possible relevance of miR-526b-3p in glioma, we assessed the expression of miR-526b-3p in glioma cell lines by RT-qPCR. As shown in Figure 1A, we found that the expression of miR-526b-3p was significantly lower in the glioma tissues compared with matched normal tissues (P<0.01). In addition, we also observed that decreased expression of miR-526b-3p was found in five glioma cell lines compared with the NHAs (Figure 1B). Thus, downregulation of miR-526b-3p may be involved in the tumorigenesis and progression of glioma. The U251 and LN229 cell line was selected for the subsequent experiments because its expression of miR-526b-3p was the lowest among these five glioma cell lines.
Relationship between miR-526b-3p expression and prognosis of glioma patients
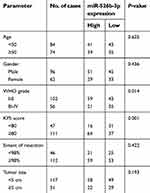
Then, we further explored the clinical significance of miR-526b-3p in glioma patients. The median expression level of miR-526b-3p was used as a cutoff point to divide all 158 patients into two groups (High and Low). As shown in Table 2, the result of chi-square analysis indicated that low miR-526b-3p expression was significantly associated with advanced WHO grade (P=0.014) and lower KPS score (P=0.001). However, no significant difference in miR-526b-3p expression was observed with age, gender, extent of resection, and tumor size (All P>0.05, Table 2). Moreover, we performed Kaplan–Meier analysis to analyzing 5-year clinical data, finding that patients with high levels of miR-526b-3p expression showed reduced overall survival (OS) time (P=0.0096, Figure 1C) and progression-free survival (PFS) time (P=0.0007, Figure 1D) compared with patients with low levels of miR-526b-3p. Subsequently, multivariate analysis demonstrated that the decreased expression of miR-526b-3p was an independent indicator for OS and PFS in glioma patients (Table 3). Taken together, our results indicated that decreased miR-526b-3p was involved in glioma development and could be identified as a poor prognostic biomarker in glioma patients.
 | Table 3 Multivariate analysis of parameters associated with overall survival and progression-free survival of glioma patients |
Forced expression of miR-526b-3p suppressed the proliferation of glioblastoma cells and induced cell apoptosis
To investigate the functional relevance of miR-526b-3p in glioblastoma cells, U251 and LN299 cells were selected for gain of function studies by transfection of miR-526b-3p mimic, because the expression levels of miR-526b-3p were significantly lower in U251 and LN299 cells than that in the other glioma cell lines. Hence, the miR-526b-3p mimic or NC mimic was transfected into U251 or LN299 cells. The efficacy of transfection was confirmed by qRT-PCR assays, and the relative expression of miR-526b-3p was increased about five times in U251 and LN299 cells after transfection of miR-526b-3p mimics (Figure 2A). The CCK-8 assays demonstrated that enhancing the expression of miR-526b-3p remarkably decreased cell viability of U251 and LN299 cells (Figure 2B and C). Moreover, colony formation assays revealed that forced expression of miR-526b-3p reduced the cell colony numbers of U251 and LN299 by about 70% (Figure 2D and E). In addition, results of flow cytometry analysis confirmed that the apoptotic rates of U251 or LN299 cells were significantly increased when they were transfected with miR-526b-3p mimics (Figure 2F and G). Collectively, these gain of function results revealed that miR-526b-3p played essential roles in the development of glioblastoma.
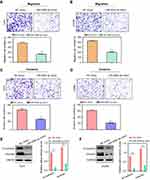
Ectopic expression of miR-526b-3p impeded the migration and invasion of glioblastoma cells
We next performed transwell assays to evaluate the effects of miR-526b-3p on the migratory and invasive capabilities of U251 or LN299 cells. The data of transwell migration assays suggested that transfection of miR-526b-3p resulted in a significant reduction (more than 50%) of migratory U251 or LN299 cells compared with the control cells (Figure 3A and B). Furthermore, the results of transwell invasion assays revealed that the upregulation of miR-526b-3p remarkably attenuated the invasive abilities of U251 and LN299 cells (Figure 3C and D). Thereafter, we assessed the protein levels of epithelial to mesenchymal related molecules by the use of western blot. The results indicated that N-cadherin as well as vimentin in U251 and LN299 cells were remarkably decreased after transfecting with miR-526b-3p mimic (Figure 3E and F). Overall, our data suggested that enhancing the expression of miR-526b-3p impaired the metastasis potentials of glioblastoma cells.
miR-526b-3p modulated the expression of WEE1 by directly targeting its 3ʹ-UTR
We next aimed to ascertain the underlying molecular mechanisms by which miR-526b-3p exerted its tumor-suppressing roles in glioblastoma. By using an online bioinformatics database (starBase,
Overexpression of WEE1 reversed the inhibitory effects of miR-526b-3p on glioblastoma cell proliferation and invasion
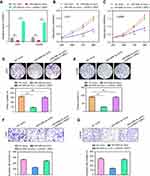
Next, we explored whether WEE1 mediated the effects of miR-526b-3p on the development and progression of glioblastoma. To achieve that, rescue experiments were carried out by co-transfecting the miR-526b-3p mimic with or without pcDNA3.1-WEE1 plasmid. The qRT-PCR assays showed that overexpression of miR-526b-3p reduced the WEE1 mRNA levels, while transfection of pcDNA3.1-WEE1 plasmid abrogated the inhibitory effects of miR-526b-3p on WEE1 expression (Figure 5A). We next performed CCK-8 assays, and the data suggested that ectopic expression of miR-526b-3p resulted in a significant reduction of proliferation, on the contrary, co-transfection of miR-526b-3p as well as pcDNA3.1-WEE1 plasmid, reversed the proliferation of U251 and LN299 cells (Figure 5B and C). Besides, re-introduction of WEE1 notably increased the number of cell colonies compared with transfection of miR-526b-3p mimic alone (Figure 5D and E). In addition, transwell invasion assays revealed that restored WEE1 expression by transfecting with pcDNA3.1-WEE1 plasmid remarkably rescued the suppressive effects of miR-526b-3p on the invasive abilities of U251 and LN299 cells (Figure 5F and G). Taken together, our data provided evidence that WEE1 was a downstream functional effector of miR-526b-3p in glioblastoma cells and miR-526b-3p modulated the development and progression of glioblastoma via WEE1.
Discussion
In China, glioblastoma multiforme is the most aggressive form with very poor survival rates.17 To better understand and to find more effective treatments for this disease, there is an urgent need to identify valuable diagnostic and prognostic biomarkers.18,19 In the past few decades, although various molecular biomarkers were identified, few of them were used in clinical practice due to the relatively low susceptibility.20,21 Recently, the potential of miRNAs as novel clinical biomarkers attracted scientists’ attention.22 In this study, we forced on a multiple functional miRNA named with miR-526b-3p.
In this study, we first detected the expression levels of miR-526b-3p in glioma patients, finding that miR-526b-3p expression was significantly reduced in both glioma tissues and cell lines. Then, chi-square test by analyzing clinical samples showed that low miR-526b-3p expression was significantly associated with advanced WHO grade and lower KPS score, which suggested that miR-526b-3p played a negative role in clinical progression of glioma. Moreover, with 5-year follow-up, clinical data were collected. Then, we performed Kaplan–Meier analysis to explore the influence of miR-526b-3p on long-term survival of glioma patients, finding that lower miR-526b-3p expression was significantly associated with poorer OS and PFS. Finally, in the Cox multivariate analysis, we confirmed that low miR-526b-3p expression was an independent poor prognostic factor for both 5-year OS and 5-year PFS. Thus, our results provided first evidence that miR-526b-3p may be used as a potential prognostic biomarker. However, the sample size was relatively small; we will collect more samples for research in the future.
MicroRNAs can have large-scale effects by modulating the expression of various genes during mammalian development and carcinogenesis.23 Recently, multiple reports indicated that miR-526b-3p has been shown to be one of the important determinants in cancers. For instance, Liu et al24 reported that miR-526b-3p was lowly expressed in hepatocellular carcinoma patients and its low levels were associated with advanced clinical progression, such as tumor size, venous infiltration, and TNM stage. In addition, in their functional experiments, it was found that forced miR-526b-3p expression significantly suppressed inhibited proliferation, migration, and invasion of hepatocellular carcinoma cells by targeting Sirtuin 7. On the other hand, downregulation of miR-526b-3p and its tumor suppressor effects were also observed in several other tumors, such as colon cancer and non-small cell lung cancer.25,26 However, the function of miR-526b-3p in glioma has not been investigated. In this study, because miR-526b-3p displayed a low level in glioma, we used miR-526b-3p mimics to upregulate the expression of miR-526b-3p and performed a series of cells experiments to study its biological function. We found that overexpression of miR-526b-3p significantly suppressed glioma cells proliferation, migration, and invasion, and promoted apoptosis. Taken together with previous reports, it suggests that miR-526b-3p functioned mainly as a tumor suppressor in glioma.
WEE1, a member of the tyrosine kinase family, is a G2/M checkpoint regulatory protein involved in the regulation of cell cycle progression.27 Massive studies reported that WEE1 expression was upregulated in various tumors, and its oncogenic roles were also widely reported, which suggested that WEE1 may be a good therapeutic candidate target in cancer treatment.28–30 In glioma, several studies reported that WEE1 was overexpressed in glioma and promoted glioma cells proliferation, migration, and invasion.31,32 However, its potential mechanism in glioma progression remains largely unclear. In this study, in order to explore the potential mechanism by which miR-526b-3p displayed its tumor suppressor role in glioma, our attention focused on the association between miR-526b-3p and WEE1. As predicted by TargetScanHuman 7.0, WEE1 mRNA was one target of miR-526b-3p. Transfection with miR-526b-3p mimic decreased the luciferase activity and protein expression of WEE1 in glioma cells, suggesting that WEE1 may be the target gene of miR-526b-3p. Moreover, we confirmed that the ectopic expression of WEE1 reversed the inhibitory effect of miR-526b-3p overexpression on cell proliferation, migration, and invasion in glioma cells, indicating that miR-526b-3p served as a tumor suppressor through WEE1. However, functional mechanisms are not deeply researched in this article. In the future, we plan to more fully describe the mechanisms and relationships with miR-526b-3p related to tumor diseases and glioma biomarkers.
Conclusion
miR-526b-3p correlates with a poor prognosis of glioma patients by regulating glioma cell proliferation, migration, and invasion via targeting WEE1. It can be a useful target and potential prognostic biomarker for glioma patients.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (Grant No. 81472594, Grant No. 81770781, and Grant No. 81560414).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Jiang T, Mao Y, Ma W, et al. Chinese Glioma Cooperative Group (CGCG). CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016 Jun 1;375(2):263–273.
4. Champ CE, Palmer JD, Volek JS, et al. Targeting metabolism with a ketogenic diet during the treatment of glioblastoma multiforme. J Neurooncol. 2014;117(1):125–131. doi:10.1007/s11060-014-1362-0
5. Ikeda H, Shiku H Antigen-receptor gene-modified T cells for treatment of glioma. Adv Exp Med Biol. 2012;746:202–215. doi:10.1007/978-1-4614-3146-6_16
6. Kang JH, Adamson C Novel chemotherapeutics and other therapies for treating high-grade glioma. Expert Opin Investig Drugs. 2015;24(10):1361–1379. doi:10.1517/13543784.2015.1048332
7. Hammond SM An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi:10.1016/j.addr.2015.05.001
8. Bak RO, Mikkelsen JG miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA. 2014;5(3):317–333. doi:10.1002/wrna.1213
9. Sand M The pathway of miRNA maturation. Methods Mol Biol. 2014;1095:3–10. doi:10.1007/978-1-62703-703-7_1
10. Gao Y, Lin L, Lin T, et al. The role of miRNA-233 in cancer: Function, diagnosis and therapy. Gene. 2017 Jun 15;616:1–7.
11. Li J, Chen Y, Guo X, et al. GPC1 exosome and its regulatory miRNAs are specific markers for the detection and target therapy of colorectal cancer. J Cell Mol Med. 2017 May; 21(5):838–847.
12. Zhang H, Wang W, Fan S. The role of miRNAs in resistance to targeted treatments of non-small cell lung cancer. Cancer Chemother Pharmacol. 2017 Feb; 79(2):227–231.
13. Di Leva G, Garofalo M, Croce CM MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi:10.1146/annurev-pathol-012513-104715
14. Bai QL, Hu CW, Wang XR, et al. MiR-616 promotes proliferation and inhibits apoptosis in glioma cells by suppressing expression of SOX7 via the Wnt signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(24):5630–5637. doi:10.26355/eurrev_201712_14006
15. Han L, Liu S, Liang J, et al. A genetic polymorphism at miR-526b binding-site in the lincRNA-NR_024015 exon confers risk of esophageal squamous cell carcinoma in a population of North China. Mol Carcinog. 2017;56(3):960–971. doi:10.1002/mc.22549
16. Kim KH, Jung JY, Son ED, et al. miR-526b targets 3ʹ UTR of MMP1 mRNA. Exp Mol Med. 2015;47:e178. doi:10.1038/emm.2015.52
17. Yang P, Wang Y, Peng X, et al. Management and survival rates in patients with glioma in China (2004–2010): a retrospective study from a single-institution. J Neurooncol. 2013;113(2):259–266. doi:10.1007/s11060-013-1103-9
18. Wick W, Weller M, van Den Bent M, et al. MGMT testing: the challenges for biomarker-based glioma treatment. Nat Rev Neurol. 2014;10(7):372–385. doi:10.1038/nrneurol.2014.100
19. Peng G, Liao Y, Shen C.miRNA-429 Inhibits astrocytoma proliferation and invasion by targeting BMI1. Pathol Oncol Res. 2017 Apr;23(2):369. doi:10.1093/bja/aeu467376.
20. Cohen AL, Colman H Glioma biology and molecular markers. Cancer Treat Res. 2015;163:15–30. doi:10.1007/978-3-319-12048-5_2
21. Appin CL, Brat DJ Biomarker-driven diagnosis of diffuse gliomas. Mol Aspects Med. 2015;45:87–96. doi:10.1016/j.mam.2015.05.002
22. Hayes J, Peruzzi PP, Lawler S MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. doi:10.1016/j.molmed.2014.06.005
23. Tutar Y miRNA and cancer: computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
24. Liu X, Yang L, Tu J, et al. microRNA-526b servers as a prognostic factor and exhibits tumor suppressive property by targeting Sirtuin 7 in hepatocellular carcinoma. Oncotarget. 2017;8(50):87737–87749. doi:10.18632/oncotarget.21209
25. Zhang R, Zhao J, Xu J, et al. miR-526b-3p functions as a tumor suppressor in colon cancer by regulating HIF-1alpha. Am J Transl Res. 2016;8(6):2783–2789.
26. Zhang ZY, Fu SL, Xu SQ, et al. By downregulating Ku80, hsa-miR-526b suppresses non-small cell lung cancer. Oncotarget. 2015;6(3):1462–1477. doi:10.18632/oncotarget.2808
27. Do K, Doroshow JH, Kummar S Wee1 kinase as a target for cancer therapy. Cell Cycle. 2013;12(19):3159–3164. doi:10.4161/cc.26062
28. Slipicevic A, Holth A, Hellesylt E, et al. Wee1 is a novel independent prognostic marker of poor survival in post-chemotherapy ovarian carcinoma effusions. Gynecol Oncol. 2014;135(1):118–124. doi:10.1016/j.ygyno.2014.07.102
29. Brockway S, Zeleznik-Le NJ WEE1 is a validated target of the microRNA miR-17-92 cluster in leukemia. Cancer Genet. 2015;208(5):279–287. doi:10.1016/j.cancergen.2015.01.001
30. Chang Q, Chandrashekhar M, Ketela T, et al. Cytokinetic effects of Wee1 disruption in pancreatic cancer. Cell Cycle. 2016;15(4):593–604. doi:10.1080/15384101.2016.1138188
31. Music D, Dahlrot RH, Hermansen SK, et al. Expression and prognostic value of the WEE1 kinase in gliomas. J Neurooncol. 2016;127(2):381–389. doi:10.1007/s11060-015-2050-4
32. Caretti V, Hiddingh L, Lagerweij T, et al. WEE1 kinase inhibition enhances the radiation response of diffuse intrinsic pontine gliomas. Mol Cancer Ther. 2013;12(2):141–150. doi:10.1158/1535-7163.MCT-12-0735
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.