Back to Journals » OncoTargets and Therapy » Volume 13
miR-525-5p Modulates Proliferation and Epithelial–Mesenchymal Transition of Glioma by Targeting Stat-1
Authors Xie P, Han Q , Liu D, Yao D, Lu X, Wang Z, Zuo X
Received 12 April 2020
Accepted for publication 23 August 2020
Published 7 October 2020 Volume 2020:13 Pages 9957—9966
DOI https://doi.org/10.2147/OTT.S257951
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Peng Xie,1,* Qiu Han,2,* Dachao Liu,3,* Dan Yao,4 Xiaoqing Lu,5 Ziyu Wang,6 Xiaohua Zuo7
1Department of Neurosurgery, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China; 2Department of Neurology, Huai’an First People’s Hospital, The Affiliated Huai’an No.1 People’s Hospital of Nanjing Medical University, Huai’an, Jiangsu 223000, People’s Republic of China; 3Department of Image, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China; 4Department of General Surgery, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China; 5Department of Orthopedic, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China; 6Department of Emergency Intensive Care Unit, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China; 7Department of Pain Management, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, Huai’an 223002, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaohua Zuo
Department of Pain Management, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, No. 62, Huaihai Road (S.), Huai’an 223002, People’s Republic of China
Email [email protected]
Ziyu Wang
Department of Emergency Intensive Care Unit, The Affiliated Huai’an Hospital of Xuzhou Medical University, The Second People’s Hospital of Huai’an, No. 62, Huaihai Road (S.), Huai’an 223002, People’s Republic of China
Email [email protected]
Background: Glioma is the most aggressive human brain tumor. Recent studies revealed that microRNAs play vital roles in glioma. However, the function of microRNA-525-5p (miR-525-5p) in glioma remains unclear.
Methods: qRT-PCR and Western blotting were used to evaluate mRNA and protein levels in glioma tissues and cells. Colony formation, CCK-8, and Edu assays evaluated the growth of glioma cells. Wound-healing, transwell, and 3D invasion assays examined the migration and invasion activities of glioma cells. Luciferase reporter assays assessed the regulatory relationship interaction between miR-525-5p and Stat-1. A mouse xenograft model was used to examine the effect of miR-525-5p on glioma in vivo.
Results: miR-525-5p expression was downregulated in glioma tissues and cells. Overexpressing miR-525-5p decreased the growth of glioma cells and reduced the migration, invasion, and epithelial–mesenchymal transition of glioma cells. Bioinformatics analysis identified Stat-1 as a potential target of miR-525-5p, and dual luciferase reporter assays revealed that miR-525-5p negatively regulates Stat-1. Decreased Stat-1 led to the inhibition of FOXM1, affecting NF-κB signaling activity. Overexpressing miR-525-5p reduced tumor development in vivo.
Conclusion: miR-525-5p negatively regulates cell proliferation, migration, invasion, and epithelial–mesenchymal transition in glioma, and Stat 1 is a target of miR-525-5p. miR-525-5p may be a potential target for glioma treatment.
Keywords: miR-525-5p, glioma, Stat 1, proliferation, EMT
Introduction
Glioma is the most aggressive human brain tumor, with high morbidity and mortality rates.1,2 Despite advances in the treatments for glioma, the prognosis is still poor.3 Therefore, it is vital to understand the mechanisms of the development of glioma, and to develop more effective therapeutic strategies.
MicroRNAs (miRNAs) are small noncoding RNAs 18 to 24 nucleotides in length that specifically bind to the 3′-untranslated region (3′- UTR) of target mRNAs.4–6 miRNAs regulate gene expression through post-transcriptional degradation of mRNA to degradation or by otherwise leading mRNA to degradation.7,8 Aberrant expression of miRNAs is associated with different kinds of tumors, including glioma.9,10
Stat-1 promotes growth of tumor cells.11 Activation of Stat family can promote tumor invasion, angiogenesis, survival and proliferation.12–14 A previous study reported that Stat-1 induced poor survival in glioblastoma.15 Furthermore, inhibition of Stat-1 induced a mesenchymal phenotype and angiogenic factors above basal levels.16
Here, we showed that microRNA-525-5p (miR-525-5p) expression was decreased in glioma cells and tissues. We found that miR-525-5p plays a vital role in regulating the proliferation, migration, invasion, and EMT of glioma cells via targeting Stat-1. Furthermore, decreasing Stat-1 led to the inhibition of FOXM1, thus affecting NF-κB signaling activity. These findings indicate that miR-525-5p could be a promising therapeutic target for glioma.
Materials and Methods
Glioma Cells and Tissues
Glioma cell lines (U87, A172, U251 and T98) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Normal human astrocytes (NHAs) were purchased from Lonza (Walkersville, MD, USA). Cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) or 1640 medium supplemented with 10% FBS (Invitrogen) in a humidified 37 °C incubator with 5% CO2.
A total of 26 nontumorous brain tissues (NBTs, obtained from the cortex of decompressive surgery patients with brain trauma or hypertensive intracerebral hemorrhage) and glioma tissues were obtained from the Department of Neurosurgery, Huai’an Hospital Affiliated to Xuzhou Medical University, Second People’s Hospital of Huai’an City. Of the 26 samples, 5 were NBTs, 9 were low-grade glioma (LGG) samples (grade II) and 12 were high-grade glioma (HGG) samples (grade III and IV). All patients were informed of the purpose of this study and signed written informed consent. This study was approved by the Ethics Committee of Xuzhou Medical University. miRNA expression data were obtained from the CGGA database (http://www.cgga.org.cn/portal.php), and gene expression data were obtained from the TCGA database (http://tcga-data.nci.nih.gov).
qRT-PCR
qRT-PCR was performed as previously described.17
Lentivirus and Cell Transfection
T98 and U251 cells were infected with lentiviruses carrying miR-525-5p or control, which were obtained from Genepharma (Shanghai, China). To overexpress Stat-1, T98 and U251 cells were transfected with Stat 1 over-expressing recombinant vector (GenePharma). After 48 h, cells were used for functional and mechanism assays.
Cell Growth Assays
Colony formation, CCK-8, and 5-ethynyl-2′-deoxyuridine (Edu) assays were performed as described previously.18
Flow Cytometry
Glioma cells were harvested, fixed with 70% alcohol and stored at −20 °C for several days. Cells were washed with PBS, treated with 50 mg/mL of propidium iodide and analyzed by flow cytometry.
Migration and Invasion Assays
For migration assays, infected cells were seeded in 6-well plates. A linear scratch was created in the cell monolayer using a pipette tip and then plates were cultured in an incubator. The cells were photographed under a microscope at 0 h and 48 h.
For invasion assays, cells were seeded in the upper chambers of Transwell plates pre-coated with 10 μg of Matrigel diluted with serum-free DMEM medium; medium with 10% FBS was added to the lower chambers. After incubation for 36 h, cells were fixed with 4% formaldehyde, stained with 1% crystal violet and photographed using a microscope (200×).
3D invasion assay was performed as described previously.19
Luciferase Reporter Assay
The regulatory effects of miR-525-5p on Stat-1 were investigated by luciferase reporter assays. Wild-type and mutant 3′-UTR sequences of Stat-1 were cloned into the pGL-3M recombinant plasmid. U251 and T98 cells overexpressing miR-525-5p or control were transfected with the luciferase reporters using Lipofectamine 3000. A plasmid expressing Renilla luciferase was co-transfected for normalization.
Subcutaneous Xenografts Models
Male nude mice were purchased from Beijing Vital River Laboratory Animal Technology. Fourteen mice were randomly assigned into two groups. U251 cells overexpressing miR-525-5p or control were harvested and resuspended in PBS. Cells (1×106) were then inoculated into nude mice. Tumor growth was monitored and tumor volume was calculated using the formula: V = 0.5×length×width2. At 30 days after injection, the mice were sacrificed and the tumors were harvested. The study has been approved by the Ethics Committee of Huai’an Hospital Affiliated to Xuzhou Medical University, Second People’s Hospital of Huai’an City, and is based on the National Institutes of Health’s Guide to the Care and Use of Laboratory Animals.
Western Blotting Assay
Western blot analysis was performed as described previously. Primary antibodies against cyclin D1 (ab40754), CDK2 (ab32147), CDK2 (ab108357), Stat1 (ab30645), Fibronectin (ab2413, Abcam); p65 (#8242), p-p65 (#3033), E-cadherin (#3195), FOXM1 (#5436), N-cadherin (#13116), Vimentin (#5741, Cell Signaling Technology), and actin (Bethyl Laboratories) were used for Western blotting analysis.
Statistical Analysis
All experiments were performed at least three times. Statistical analyses were performed with GraphPad Prism 5. The results are expressed as mean ± SD. Statistical differences were analyzed by Student’s t-test or χ2 test. P<0.05 was considered to indicate statistical significance.
Results
MiR-525-5p Expression is Decreased in Glioma Cells and Tissues
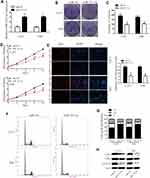
We first analyzed the expression of miR-525-5p in the CGGA database. We found that miR-525-5p expression level was higher in LGGs compared with that in HGGs (Figure 1A). We next detected miR-525-5p expression in 26 samples (5 NBTs, 9 LGGs, and 12 HGGs) via qRT-PCR. miR-525-5p level was decreased in LGGs and HGGs compared with NBTs (Figure 1B). The level of miR-525-5p was markedly decreased to 56.6% in LGGs and 18.9% in HGGs compared with levels in NBTs. We then investigated miR-525-5p expression in glioma cell lines (U87, A172, U251, and T98) and NHAs. miR-525-5p expression was decreased in glioma cells compared with NHAs: miR-525-5p level was decreased to 60.6% in U87 cells, 59.0% in A172 cells, 37.3% in U251 cells, and 32.0% in T98 cells (Figure 1C).
Overexpressing miR-525-5p Decreases the Growth of Glioma Cells
We transfected miR-525-5p or the negative control into U251 and T98 glioma cells and confirmed miR-525-5p expression in the cell lines by qRT-PCR (Figure 2A). To determine the effect of miR-525-5p on the growth of glioma cells, we performed colony formation, CCK8, and Edu assays. Colony formation assays indicated that overexpressing miR-525-5p decreased the proliferation of U251 and T98 cells compared with controls (Figure 2B and C). CCK8 assay showed that overexpressing miR-525-5p inhibited the growth of both U251 and T98 cells (Figure 2D). EdU revealed similar results (Figure 2E).
We further found that overexpressing miR-525-5p induced cell cycle arrest in glioma cells at G1 (Figure 2F and G). The altered expressions of cell cycle-related proteins CDK2, CDK4 and cyclin D1 were consistent with this result (Figure 2H). These results indicate that overexpression of miR-525-5p decreases the proliferation of glioma cells and inducing a G1 cell cycle arrest.
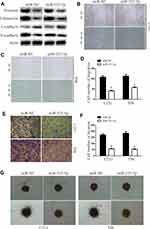
Overexpressing miR-525-5p Reduces EMT in Glioma Cells
We next examined the effects of miR-525-5p on EMT. The levels of mesenchymal markers vimentin, fibronectin, and N-cadherin were downregulated, while E-cadherin levels were upregulated in U251 and T98 cells overexpressing miR-525-5p (Figure 3A). Wound healing assays demonstrated that overexpressing miR-525-5p reduced the migration ability of glioma cells (Figure 3B–D). Furthermore, transwell and 3D-invasion assays showed that overexpressing miR-525-5p reduced the invasion ability of glioma cells (Figure 3E–G). Together these results indicate that overexpression of miR-525-5p reduces EMT, migration, and invasion in glioma cells.
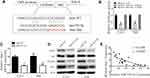
Stat 1 is a Direct Target of miR-525-5p
To search for the molecular mechanism underlying the inhibitory effect of miR-525-5p on proliferation, migration, invasion, and EMT in glioma, we used TargetScan and miRanda algorithms to identify potential targets of miR-525-5p.20,21 Bioinformatics analysis identified binding sites for miR-525-5p in Stat 1 mRNA (Figure 4A). Luciferase reporter assay was used to examine the effect of miR-525-5p on Stat-1. Decreased luciferase activity of reporter vector containing the wild-type 3′-UTR of Stat-1 was observed after transfection with miR-525-5p, whereas no effects were observed with the mutant 3′-UTR reporter vector in which the putative miR-525-5p binding sites were mutated (Figure 4B). We further found that miR-525-5p downregulated the mRNA and protein levels of Stat-1 (Figure 4C and D). Reduction of Stat-1 led to the inhibition of FOXM1, affecting NF-κB signaling activity (Figure 4D). Pearson’s correlation analysis showed a negative correlation between miR-525-5p and Stat-1 in glioma (Figure 4E).
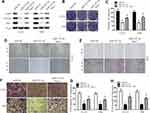
Stat 1 Attenuates the Effects of miR-525-5p on Glioma Cell Proliferation, Migration, Invasion, and EMT
We co-overexpressed Stat-1 together with miR-525-5p in U251 and T98 cells and confirmed that co-overexpressing Stat-1 restored the miR-525-5p-mediated effects on Stat-1 protein levels in U251 and T98 cells (Figure 5A). Cell proliferation, migration, and invasion assays showed that co-overexpressing Stat-1 attenuated the inhibitory effects of miR-525-5p on glioma proliferation, migration, and invasion (Figure 5B–H). These results indicated that Stat-1 is a functional target of miR-525-5p in glioma.
miR-525-5p Exhibits a Tumor Suppressor Role in vivo
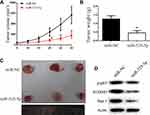
We next explored the function of miR-525-5p in glioma development in vivo using subcutaneous xenografts. Nude mice were inoculated with U251 cells overexpressing control miRNA or miR-525-5p. Mice injected with cells overexpressing miR-525-5p showed inhibited tumor growth in vivo compared with controls (Figure 6A–C). We also observed downregulated protein levels of Stat-1, FOXM1, and p-p65 in tumors derived from miR-525-5p-overexpressing cells (Figure 6D), which is consistent with the in vitro results. Taken together, our results suggest that miR-525-5p inhibits the development of glioma both in vitro and in vivo.
Discussion
miRNAs play important roles in the development of cancers, including glioma, and can function as tumor suppressors or oncogenes by regulating the target genes.22,23 Therefore, miRNAs may be potential diagnostic and therapeutic targets in glioma.24
Dysregulated miRNAs are important for the pathology of glioma.25,26 Here, we found that miR-525-5p was downregulated in both glioma tissues and human glioma cells. We observed a significant decrease of glioma cell growth when miR-525-5p was overexpressed, and overexpressing miR-525-5p induced cell cycle arrest in G1. We further showed that miR-525-5p promoted EMT, migration, and invasion of glioma cells. Additionally, we confirmed a tumor inhibitory role of miR-525-5p in a xenograft tumor model.
Stat-1 closely involved in several biological functions, including cell proliferation, differentiation, migration, invasion, and apoptosis.27 Previous studies have reported that Stat-1 is upregulated in different types of tumors, including glioma.28–30 Stat-1 is regulated by IGFBP-3 and is a predictor for poor survival in glioblastoma.31 Moreover, Stat-1 is downregulated in gastric cancer tissue and involved in cell metastasis.32 We investigated the mechanisms underlying the inhibitory effect of miR-525-5p on glioma cell proliferation and EMT using bioinformatics analysis, and the results identified Stat-1 as a predicted target molecule of miR-525-5p. Overexpressing miR-525-5p reduced the protein and mRNA levels of Stat-1. Stat-1 inhibited NF-κB signaling through FOXM1.28 Additionally, restoration of Stat-1 expression could rescue the inhibition of cell proliferation and EMT that resulted from miR-525-5p overexpression. Together these data indicate that Stat-1 is a direct target gene of miR-525-5p and that Stat-1 attenuates the effects of miR-525-5p on the proliferation and EMT of glioma cells.
Conclusions
In this study, we investigated the effects of miR-525-5p on glioma. miR-525-5p was downregulated in glioma cells and tissues. Functional experiments indicated that miR-525-5p inhibits the proliferation, migration, invasion, and EMT of glioma and targets Stat-1. These findings suggest that miR-525-5p may be a potential target for glioma.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author (Xiaohua Zuo) on reasonable request.
Ethics Approval and Consent to Participate
The study protocol was designed and approved by the ethical committee of the Affiliated Huai’an Hospital of Xuzhou Medical University, the Second People’s Hospital of Huai’an. All animal experiments were conducted under the approval of the Affiliated Huai’an Hospital of Xuzhou Medical University, the Second People’s Hospital of Huai’an, and is based on the National Institutes of Health’s Guide to the Care and Use of Laboratory Animals.
Consent for Publication
All authors agree on publication of the results of the present manuscript.
Acknowledgments
We thank Gabrielle White Wolf, PhD, from Liwen Bianji, Edanz Editing China, for editing the English text of a draft of this manuscript.
Author Contributions
XHZ, ZYW and PX designed the experiments; PX, QH, and DCL performed the experiments; DY and XQL analysis the results; PX wrote the paper. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Research Program of Science and Technology Support Program of Huai’an (HAB201931) and Open Project of Key Laboratory of Universities of Jiangsu Province (XZSYSKF2019002).
Disclosure
The authors declare that they have no competing interests.
References
1. da Hora CC, Pinkham K, Carvalho L, et al. Sustained NF-kappaB-STAT3 signaling promotes resistance to Smac mimetics in Glioma stem-like cells but creates a vulnerability to EZH2 inhibition. Cell Death Discov. 2019;5:72.
2. Hoeman CM, Cordero FJ, Hu G, et al. ACVR1 R206H cooperates with H3.1K27M in promoting diffuse intrinsic pontine glioma pathogenesis. Nat Commun. 2019;10(1):1023. doi:10.1038/s41467-019-08823-9
3. Phillips RE, Yang Y, Smith RC, et al. Target identification reveals lanosterol synthase as a vulnerability in glioma. Proc Natl Acad Sci U S A. 2019;116(16):7957–7962. doi:10.1073/pnas.1820989116
4. Ge J, Li J, Na S, Wang P, Zhao G, Zhang X. miR-548c-5p inhibits colorectal cancer cell proliferation by targeting PGK1. J Cell Physiol. 2019;234(10):18872–18878. doi:10.1002/jcp.28525
5. Yin J, Zeng A, Zhang Z, Shi Z, Yan W, You Y. Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine. 2019;42:238–251. doi:10.1016/j.ebiom.2019.03.016
6. Yang J, Li C, Li H. LncRNA CACNA1G-AS1 facilitates hepatocellular carcinoma progression through the miR-2392/C1orf61 pathway. J Cell Physiol. 2019;234(10):18415–18422. doi:10.1002/jcp.28477
7. Jin C, Li M, Ouyang Y, Tan Z, Jiang Y. MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J Neurooncol. 2017;133(2):247–255. doi:10.1007/s11060-017-2438-4
8. Yang JK, Yang JP, Tong J, et al. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J Neurooncol. 2017;131(2):255–265. doi:10.1007/s11060-016-2308-5
9. Chen Q, Guo W, Zhang Y, Wu Y, Xiang J. MiR-19a promotes cell proliferation and invasion by targeting RhoB in human glioma cells. Neurosci Lett. 2016;628:
10. Qi Z, Cai S, Cai J, et al. miR-491 regulates glioma cells proliferation by targeting TRIM28 in vitro. BMC Neurol. 2016;16(1):248.
11. Sasidharan Nair V, Toor SM, Ali BR, Elkord E. Dual inhibition of STAT1 and STAT3 activation downregulates expression of PD-L1 in human breast cancer cells. Expert Opin Ther Targets. 2018;22(6):547–557. doi:10.1080/14728222.2018.1471137
12. Zhou W, Grandis JR, Wells A. STAT3 is required but not sufficient for EGF receptor-mediated migration and invasion of human prostate carcinoma cell lines. Br J Cancer. 2006;95(2):164–171. doi:10.1038/sj.bjc.6603234
13. Zhang N, Duan WD, Leng JJ, et al. STAT3 regulates the migration and invasion of a stemlike subpopulation through microRNA21 and multiple targets in hepatocellular carcinoma. Oncol Rep. 2015;33(3):1493–1498.
14. Han Y, Chen M, Wang A, Fan X. STAT3-induced upregulation of lncRNA CASC11 promotes the cell migration, invasion and epithelial-mesenchymal transition in hepatocellular carcinoma by epigenetically silencing PTEN and activating PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2019;508(2):472–479. doi:10.1016/j.bbrc.2018.11.092
15. Duarte CW, Willey CD, Zhi D, et al. Expression signature of IFN/STAT1 signaling genes predicts poor survival outcome in glioblastoma multiforme in a subtype-specific manner. PLoS One. 2012;7(1):e29653. doi:10.1371/journal.pone.0029653
16. Arslan AD, Sassano A, Saleiro D, et al. Human SLFN5 is a transcriptional co-repressor of STAT1-mediated interferon responses and promotes the malignant phenotype in glioblastoma. Oncogene. 2017;36(43):6006–6019. doi:10.1038/onc.2017.205
17. Du X, Tu Y, Liu S, et al. LINC00511 contributes to glioblastoma tumorigenesis and epithelial-mesenchymal transition via LINC00511/miR-524-5p/YB1/ZEB1 positive feedback loop. J Cell Mol Med. 2020;24(2):1474–1487. doi:10.1111/jcmm.14829
18. Tu Y, Niu M, Xie P, et al. Smoothened is a poor prognosis factor and a potential therapeutic target in glioma. Sci Rep. 2017;7:42630.
19. Zeng A, Yin J, Li Y, et al. miR-129-5p targets Wnt5a to block PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death Dis. 2018;9(3):394. doi:10.1038/s41419-018-0343-1
20. Xu X, Bao Z, Liu Y, Ji J, Liu N. MicroRNA-98 attenuates cell migration and invasion in glioma by directly targeting Pre-B cell leukemia Homeobox 3. Cell Mol Neurobiol. 2017;37(8):1359–1371.
21. Xu X, Cai N, Zhi T, et al. MicroRNA-1179 inhibits glioblastoma cell proliferation and cell cycle progression via directly targeting E2F transcription factor 5. Am J Cancer Res. 2017;7(8):1680–1692.
22. Rani SB, Rathod SS, Karthik S, Kaur N, Muzumdar D, Shiras AS. MiR-145 functions as a tumor-suppressive RNA by targeting Sox9 and adducin 3 in human glioma cells. Neuro Oncol. 2013;15(10):1302–1316. doi:10.1093/neuonc/not090
23. Shi Z, Chen Q, Li C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 2014;16(10):1341–1353.
24. Wang Z, Wang B, Shi Y, et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34(11):1407–1419. doi:10.1038/onc.2014.75
25. Wang J, Sai K, Chen F-R, Chen Z-P. miR-181b modulates glioma cell sensitivity to temozolomide by targeting MEK1. Cancer Chemother Pharmacol. 2013;72(1):147–158. doi:10.1007/s00280-013-2180-3
26. Wei J, Wang F, Kong LY, et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73(13):3913–3926. doi:10.1158/0008-5472.CAN-12-4318
27. Jin Y, Zhou T, Li N, et al. JAK and STAT members in channel catfish: identification, phylogenetic analysis and expression profiling after Edwardsiella ictaluri infection. Dev Comp Immunol. 2018;81:
28. Liu C, Shi J, Li Q, et al. STAT1-mediated inhibition of FOXM1 enhances gemcitabine sensitivity in pancreatic cancer. Clin Sci (Lond). 2019;133(5):645–663. doi:10.1042/CS20180816
29. Laimer K, Spizzo G, Obrist P, et al. STAT1 activation in squamous cell cancer of the oral cavity: a potential predictive marker of response to adjuvant chemotherapy. Cancer. 2007;110(2):326–333. doi:10.1002/cncr.22813
30. Chen J, Zhao J, Chen L, et al. STAT1 modification improves therapeutic effects of interferons on lung cancer cells. J Transl Med. 2015;13:293.
31. Thota B, Arimappamagan A, Kandavel T, et al. STAT-1 expression is regulated by IGFBP-3 in malignant glioma cells and is a strong predictor of poor survival in patients with glioblastoma. J Neurosurg. 2014;121(2):374–383. doi:10.3171/2014.4.JNS131198
32. Chen P, Zhao D, Sun Y, Huang L, Zhang S, Yuan Y. Protein inhibitor of activated STAT-1 is downregulated in gastric cancer tissue and involved in cell metastasis. Oncol Rep. 2012;28(6):2149–2155. doi:10.3892/or.2012.2030
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.