Back to Journals » OncoTargets and Therapy » Volume 12
miR-520b Promotes Breast Cancer Stemness Through Hippo/YAP Signaling Pathway
Authors Zhang H, Lang T , Zou D, Zhou L, Lou M, Liu J, Li Y, Ding D, Li Y , Zhang N, Zheng X, Zeng X, Zhou Q , Li L
Received 30 October 2019
Accepted for publication 14 December 2019
Published 31 December 2019 Volume 2019:12 Pages 11691—11700
DOI https://doi.org/10.2147/OTT.S236607
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Hui Zhang,1–4,* Ting-yuan Lang,5,* Dong-ling Zou,5 Lei Zhou,6–8 Meng Lou,5 Jing-shu Liu,9 Yun-zhe Li,9 Dong-yan Ding,9 Yu-cong Li,5,9 Na Zhang,5 Xiao-dong Zheng,2 Xiao-hua Zeng,2 Qi Zhou,1,3–5 Li Li1
1Department of Gynecologic Oncology, Affiliated Tumor Hospital of Guangxi Medical University, Nanning 530021, Guangxi, People’s Republic of China; 2Breast Cancer Center, Chongqing University Cancer Hospital and Chongqing Cancer Institute and Chongqing Cancer Hospital, Chongqing 400030, Chongqing, People’s Republic of China; 3Chongqing Key Laboratory of Translational Research for Cancer Metastasis and Individualized Treatment, Chongqing University Cancer Hospital and Chongqing Cancer Institute and Chongqing Cancer Hospital, Chongqing 400030, Chongqing, People’s Republic of China; 4Key Laboratory for Biorheological Science and Technology of Ministry of Education (Chongqing University), Chongqing University Cancer Hospital and Chongqing Cancer Institute and Chongqing Cancer Hospital, Chongqing, Chongqing, 400030, People’s Republic of China; 5Department of Gynecologic Oncology, Chongqing University Cancer Hospital and Chongqing Cancer Institute and Chongqing Cancer Hospital, Chongqing 400030, Chongqing, People’s Republic of China; 6Department of Ophthalmology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 119228, Singapore; 7Singapore Eye Research Institute, Singapore 169856, Singapore; 8Ophthalmology and Visual Sciences Academic Clinical Research Program, Duke-NUS Medical School, Singapore 169867, Singapore; 9Bioengineering College of Chongqing University, Chongqing University, Chongqing 400030, Chongqing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Li Li
Department of Gynecologic Oncology, Affiliated Tumor Hospital of Guangxi Medical University, Nanning 530021, Guangxi, People’s Republic of China
Email [email protected]
Qi Zhou
Department of Gynecologic Oncology, Chongqing University Cancer Hospital and Chongqing Cancer Institute and Chongqing Cancer Hospital, Chongqing 400030, Chongqing, People’s Republic of China
Email [email protected]
Introduction: The breast cancer stem cells contribute to the initiation, progression, recurrence, metastasis as well as resistance of breast cancer. However, the mechanisms underlying the maintenance of breast cancer stemness have not been fully understood.
Materials and methods: TCGA and GEO data were used for measuring miR-520b expression in breast cancer tissues. Kaplan-meier analysis was used for determining the relationship between miR-520b expression level and the prognosis of patients. Genetic manipulation was performed by lentivirus system and miR-520b inhibitor was used for knockdown of miR-520b. qRT-PCR and Western blot were employed to determine the mRNA and protein levels, respectively. The stemness and EMT (Epithelial to mesenchymal transition) were assessed by sphere-formation and transwell assay as well as the expression of the related markers. The target genes of miR-520b were identified using the online database starBase V3.0.
Results: miR-520b is upregulated in cancer tissues of breast cancer patients and predicts poor prognosis. Upregulation of miR-520b was found in breast cancer stem cells. Ectopic expression of miR-520b promotes the stemness of the breast cancer cells, conversely, depletion of miR-520b attenuates the stemness of these cells. miR-520b positively regulates Hippo/YAP signaling pathway and overexpression of LAST2 abolished the effect of miR-520b on the stemness of breast cancer cells.
Conclusion: miR-520b promotes the stemness of breast cancer patients by activating Hippo/YAP signaling via targeting LATS2.
Keywords: miR-520b, breast cancer, stemness, Hippo/YAP, LATS2
Introduction
Breast cancer is recognized as one of the leading causes of cancer-related mortality in women worldwide.1 In spite of the development of aggressive interventions, the inevitability of relapse and metastasis result in severe mortality rate.2 While, the roots of breast cancer relapse and metastasis have remained elusive.
Cancer stem cells (CSCs) are a small but significant subpopulation of undifferentiated cells in tumor tissues.3–5 Accumulating evidences indicated that CSCs drive cancer initiation, progression, spread and resistance to chemo- or radiotherapy.6,7 CSCs were also identified in breast cancer tissues, called breast cancer stem cells (BCSCs). BCSCs were characterized with the ability of self-renewal, differentiation, tumor initiation, invasion and resistance of conventional therapy, which indicated that BCSCs should be an effective target for breast cancer therapy.8–12 However, our understanding of BCSCs still needs to be improved.
There are several signaling pathways that have been identified to be associated with the maintenance of breast cancer stemness, including Hippo/YAP, Notch, Hedgehog and Wnt pathways.13–18 In addition, numerous miRNAs are identified to be dysregulated in BCSCs population, such as miR-200c19, miR-205,20 miR-141,21 miR-1,22 miR-34a23, miR-221,24 miR-2125 and Let-7.26
miR-520b is a member of miR-302/372/373/520 family. Recently, several members in this family have been reported to be associated with cancers. For example, miR-373 and miR-520c/h have been reported with the role of oncogene in breast and esophageal cancer cells.27–29 While, miR-302, 372, 520a/b/e/h have been shown as a tumor suppressor in some types of cancers,30–34 which indicated that the function of the members of this family largely depends on the cellular context in specific tissue type.
In this study, we report that miR-520b promotes the stemness of breast cancer patients by activating Hippo/YAP signaling via targeting LATS2.
Materials and Methods
Cells, Clinical Samples and Cell Culture
MCF-7 and MDA-MB-231 breast cancer cells were purchased from Chinese Academy of Sciences (Shanghai, China). Both cells were cultured in Eagle’s Minimum Essential Medium (EMEM, Thermo Fisher, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Thermo Fisher, Waltham, MA, USA). All tissues were obtained from Chongqing University Cancer Hospital, which was approved by ethics committee of Chongqing University Cancer Hospital. All patients provided written informed consent, and that this was conducted in accordance with the Declaration of Helsinki. The clinical information of the patients was provided in Supplementary materials.
Stem Cell Isolation
The cells were digested, washed and re-seeded in stem cell medium (Thermo Fisher Scientific) in Ultra-low adherent plate (Corning) and cultured at 37°C in a 5% CO2 humidified incubator for 15–20 days.
Antibodies, Primers and Reagents
The detail information of antibodies and primers were provided in Supplementary materials. miR-520b inhibitors were purchased Sigma-Aldrich (St. Louis, MO, USA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Plasmids
Lentivirus containing miR-520b (HLMIR0696) and miR-520b inhibitor (HSTUD0696) were purchased from Sigma Aldrich Merck (St. Louis, MO, USA). The coding sequence (CDs) region of LATS2 CDS was synthesized by PCR and was cloned into pCDH plasmids for virus package.
Lentivirus Package, Transfection and Stable Cell Selection
HEK293T cells were co-transfected with reconstructed plasmids, VSV-G (envelop plasmid) and delta R8.2 (packaging plasmid) and were cultured for 7–14 days. The media were harvested for virus concentration using a 0.45 μm filter and Lenti-X Concentrator (Clontech, Mountain View, CA, USA). MCF-7 and MDA-MB-231 cells were infected with lentivirus, and two days after infection, the cells were cultured with complete media with puromycin. The stable cell lines were verified by Western blot assay.
Quantitative Real-Time Reverse-Transcription PCR
RNAzol RT reagent (Molecular Research Center, Cincinnati, OH, USA) was employed for Total RNA isolation. SuperScript III Platinum SYBR Green One-Step qRT-PCR Kit (Thermo Fisher, Waltham, MA, USA) was used for quantitative reverse transcriptase PCR (qRT-PCR) and GAPDH was used as an internal control.
Western Blot
The protein samples were fist separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto Immobilon-P membranes (Millipore-Sigma, St. Louis, MO, USA). The membranes were then incubated with the primary antibody at 4°C overnight. After incubation with relevant horseradish peroxidase (HRP)-conjugated secondary antibody, the Clarity™ Western ECL Substrate (BioRad, Hercules, CA, USA) was used to produce the signals.
Transwell Assay
About 2×103 cells in serum-free culture media were placed on the upper layer of the culture insert with permeable membrane (Thermo Fisher, Waltham, MA, USA). The complete culture medium was then added in the culture well (Thermo Fisher, Waltham, MA, USA). Following an incubation for 18 hrs, the cells that have migrated through the membrane are stained and counted.
Statistics
At least three replicates were performed for each experiment. Data are represented as mean ± s.d. and the significance was determined by Student’s t-test.
Results
miR-520b Is Upregulated in Breast Cancer Tissues, Predicts Poor Prognosis and Associates Breast Cancer Stem Cells
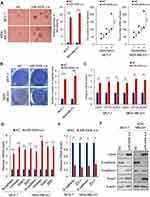
To investigate the role of miR-520b in breast cancer, we first analyzed the expression of miR-520b in breast cancer tissues. GSE57897 dataset was downloaded from GEO (Gene expression omnibus) database and we found that the expression of miR-520b was significantly upregulated in breast cancer tissues, compared with adjacent normal tissues (Figure 1A). In addition, the result from Kaplan–Meier analysis using TCGA (The cancer genome atlas) data downloaded from the Kaplan–Meier Plotter database showed that high expression of miR-520b is associated with poor prognosis of breast cancer patients (Figure 1B).
Next, we isolated BCSCs from MCF-7 and MDA-MB-231 breast cancer cells by suspension culture and analyzed the expression of miR-520b in suspension-cultured spheres and adherent or re-adherent cells by qRT-PCR. As shown in Figure 1C, the mRNA level of miR-520b is significantly upregulated in suspension-cultured spheres (MCF-7 and MDA-MD-231) compared with adherent cells and re-adherent cells, respectively. In addition, as shown in Figure 1D, the expression of miR-520b is upregulated in CD44+, CD133+, ALDH1+ MCF-7 and MDA-MD-231 cells compared with CD44-, CD133-, ALDH1- cells. Moreover, the correlation between the expression of miR-520b and CD44, CD133 and ALDH1 was analyzed by Spearman analysis. We found that the expression of miR-520b is positively correlated with CD44, CD133 and ALDH1 in breast cancer tissues (Figure 1E). The clinical information of the patients was shown in Supplementary Table 1.
These results thus suggested that miR-520b may associate with the maintenance of breast cancer stemness.
Ectopic Expression of miR-520b Promotes the Stemness of Breast Cancer Cells
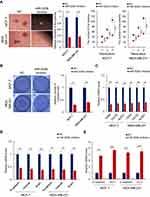
To further confirm the role of miR-520b in the maintenance of breast cancer stemness, we overexpressed miR-520b in MCF-7 and MDA-MB-231 cells (Supplementary Figure 1A). We first investigated the effect of miR-520b on the sphere-formation activities of breast cancer cells. As shown in Figure 2A, we found that miR-520b overexpression significantly increased the diameter and the number of the spheres, which demonstrated that miR-520b overexpression increased the sphere-formation activities of the breast cancer cells. As shown in Figure 2B, the results from transwell assay showed that the migration activity of the cells was significantly increased by miR-520b. In addition, the mRNA levels of the stem cell markers (CD44, CD133 and ALDH1) and mesenchymal markers (N-cadherin, Vimentin, Snail1 and ZEB1) were significantly upregulated in miR-520b-overexpressing cells (Figure 2C and D). Conversely, the mRNA levels of epithelial markers (E-cadherin and ZO-1) were significantly downregulated by miR-520b as identified by qRT-PCR (Figure 2E). The regulatory effects of miR-520b on CD44, E-cadherin, N-cadherin and Snail1 were also confirmed by Western blot assay (Figure 2F). Taken together, above results indicated that miR-520b promotes the stemness of breast cancer cells.
Knockdown of miR-520b Attenuates the Stemness of Breast Cancer Cells
Next, to confirm the stimulatory role of miR-520b in the maintenance of breast cancer stemness, miR-520b was inhibited by miR-520b inhibitor (Supplementary Figure 1B). As we expected, the sphere-formation activity of the cells was inhibited by miR-520b inhibition (Figure 3A). miR-520b inhibition also attenuated the migration activity of the breast cancer cells (Figure 3B). The mRNA levels of cancer stem cell markers and the mesenchymal markers were significantly inhibited by miR-520b inhibition in the breast cancer cells (Figure 3C and D). While, the mRNA levels of epithelial markers were significantly upregulated in miR-520b-knockdown cells (Figure 3E). These results thus confirmed the conclusion that miR-520b promotes the stemness of breast cancer cells.
miR-520b Activates Hippo/YAP Signaling in Breast Cancer Cells
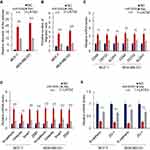
To investigate the mechanism underlying miR-520b promoting the stemness of breast cancer cells, we analyzed the potential target genes of miR-520b using the online database starBase V3.0. We found that a key component in Hippo/Yap signaling, LATS2, is the potential target of miR-520b (Figure 4A). In addition, bioinformatic analysis of GSE109169 dataset showed that the expression of LATS2 is significantly downregulated in breast cancer tissues (Supplementary Figure 1C), which indicated that inhibition of LATS2 expression may be the cause of breast cancer development. As Hippo/YAP signaling pathway is largely associated with breast cancer stemness, we next investigated whether miR-520b regulates Hippo/YAP signaling in breast cancer cells. As shown in Figure 4B, the mRNA level of LATS2 was significantly inhibited by miR-520b in breast cancer cells. In addition, the mRNA levels of YAP target genes were significantly decreased in breast cancer cells (Figure 4C). Moreover, the phosphorylation level of YAP in breast cancer cells was significantly decreased by miR-520b (Figure 4D). Furthermore, the protein level of nuclear YAP was significantly upregulated by miR-520b in breast cancer cells (Figure 4E). These results demonstrated that miR-520b activates Hippo/YAP signaling in breast cancer cells.
Overexpression of LATS2 Abolished the Effect of miR-520b on Breast Cancer Stemness
Next, we performed rescue experiment by overexpression of LATS2 in breast cancer cells (Supplementary Figure 1D) to confirm the role of Hippo/YAP signaling in the effect of LATS2 on breast cancer stemness. As shown in Figure 5A, we found that overexpression of LATS2 significantly restored the sphere-formation activity of miR-520b-overexpressing cells. Similarly, the miR-520b-induced increased migration activity of the cells was reduced by LATS2 overexpression (Figure 5B). Moreover, overexpression of LATS2 significantly abolished the effect of miR-520b on the expression of stem cell markers, mesenchymal markers as well as epithelial markers as indicated by qRT-PCR (Figure 5C–E). Thus, these results demonstrated that miR-520b promotes breast cancer stemness through Hippo/YAP signaling pathway by targeting LATS2.
Discussion
Breast cancer is one of the leading causes of tumor-related mortality worldwide.1 Recent studies indicated that BCSCs are the root of breast cancer initiation, progression, metastasis and resistance to conventional therapy.8–12 However, the mechanisms underlying the maintenance of the stemness of breast cancer cells have remained elusive. In this study, we found that miR-520b is an oncogene that promotes the stemness of BCSCs through Hippo/YAP signaling. Our finding thus improved our understanding of mechanisms underlying the maintenance of breast cancer stemness.
Through bioinformatic analysis, we first found that miR-520b is upregulated in breast cancer tissues compared with adjacent normal tissues (Figure 1A). In addition, subsequent analysis showed that upregulation of miR-520b is associated with poor prognosis of breast cancer patients (Figure 1B). Moreover, ectopic expression of miR-520b promotes the sphere-formation and migration activities (Figure 2A and B). Conversely, knockdown of miR-520b attenuates the sphere formation and migration activities (Figure 3A and B). These data thus demonstrated that miR-520b is an oncogene. Our finding is opposed to the finding of Zhang et al in which miR-520b showed as a tumor suppressor by targeting MEKK2 and cyclin D1 in hepatoma cells.32 This information indicates that the function of miR-520b and other members in this family largely depends on the cellular context and the further studies are needed for understanding of the switch of such regulation.
Furthermore, we found that the expression of miR-520b is positively correlated with stem cells markers (Figure 1E). In addition, the expression of miR-520b is upregulated in spheres and cells expressed a high level of cancer stem cell markers (CD44, CD133 and ALDH1) compared with adherent and re-adherent control cells and cancer stem cell markers negative cells (Figure 1C and D). Moreover, we found that overexpression of miR-520b promotes the stemness of cancer stem cells, including sphere-formation, migration activities as well as the expression of stem cell markers, mesenchymal markers and epithelial markers (Figure 2). Conversely, knockdown of miR-520b inhibits the stemness of breast cancer cells (Figure 3). These results thus demonstrated that miR-520b promotes the stemness of breast cancer cells.
Hippo/YAP signaling pathway plays a central role in organ size control, tissue development and stemness maintenance of both normal and cancer stem cells.35 In Hippo/YAP signaling, LATS2 is activated by MST1/2-induced phosphorylation and then phosphorylates and inhibits YAP-mediated gene transcription; most of YAP target genes positively regulates cell cycle and stemness maintenance.36,37 In this study, we found that LATS2, the potential target of miR-520b (Figure 4A), was downregulated by miR-520b overexpression (Figure 4B). Correspondingly, the target genes of Hippo/YAP signaling pathway were inhibited in miR-520b-overexpressing cells (Figure 4C). Moreover, the phosphorylation of YAP is downregulated by miR-520b overexpression (Figure 4D). Furthermore, the protein level of nuclear YAP was significantly upregulated by miR-520b (Figure 4E). These results demonstrated that Hippo/YAP signaling is positively regulated by miR-520b in breast cancer cells. Subsequent rescue experiments showed that inhibition of LATS2 is necessary for miR-520b promoting the stemness of BCSCs. Taken together, these results demonstrated that miR-520b promotes breast cancer stemness through Hippo/YAP signaling pathway by targeting LATS2.
Conclusion
In conclusion, our results demonstrated that miR-520b promotes the stemness of breast cancer cells by activating Hippo/YAP signaling pathway via targeting LATS2.
Acknowledgment
This study was supported by Chongqing Science & Technology Commission (Grant Nos. cstc2017jxjl130021, cstc2019jscx-msxmX0106, cstc2019jscx-msxmX0174, and cstc2017jxjl130047).
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers. 2019;5(1):66. doi:10.1038/s41572-019-0111-2
2. Pondé NF, Zardavas D, Piccart M. Progress in adjuvant systemic therapy for breast cancer. Nat Rev Clin Oncol. 2019;16(1):27–44. doi:10.1038/s41571-018-0089-9
3. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi:10.1038/nm.4409
4. Nguyen LV, Vanner R, Dirks P, et al. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12(2):133–143. doi:10.1038/nrc3184
5. Shibue T, RA W. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi:10.1038/nrclinonc.2017.44
6. Zardavas D, Baselga J, Piccart M. Emerging targeted agents in metastatic breast cancer. Nat Rev Clin Oncol. 2013;10(4):191–210. doi:10.1038/nrclinonc.2013.29
7. Akebe N, Harris PJ, Warren RQ, et al. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8(2):97–106. doi:10.1038/nrclinonc.2010.196
8. de Beça FF, Caetano P, Gerhard R, et al. Cancer stem cells markers CD44, CD24 and ALDH1 in breast cancer special histological types. J Clin Pathol. 2013;66:187–191. doi:10.1136/jclinpath-2012-201169
9. Koren S, Reavie L, Couto JP, et al. PIK3CAH1047R induces multipotency and multi-lineage mammary tumours. Nature. 2015;525(7567):114–118. doi:10.1038/nature14669
10. Chaffer CL, Marjanovic ND, Lee T, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154(1):61–74. doi:10.1016/j.cell.2013.06.005
11. Lagadec C, Vlashi E, Della Donna L, et al. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30(5):833–844. doi:10.1002/stem.1058
12. Pires BR, DE Amorim ÍS, Souza LD, et al. Targeting cellular signaling pathways in breast cancer stem cells and its implication for cancer treatment. Anticancer Res. 2016;36(11):5681–5691. doi:10.21873/anticancer.11151
13. D’Angelo RC, Ouzounova M, Davis A, et al. Notch reporter activity in breast cancer cell lines identifies a subset of cells with stem cell activity. Mol Cancer Ther. 2015;14:779–787. doi:10.1158/1535-7163.MCT-14-0228
14. O’Toole SA, Machalek DA, Shearer RF, et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res. 2011;71:4002–4014. doi:10.1158/0008-5472.CAN-10-3738
15. Harris LG, Pannell LK, Singh S, et al. Increased vascularity and spontaneous metastasis of breast cancer by Hedgehog signaling mediated upregulation of cyr61. Oncogene. 2012;31(28):3370–3380. doi:10.1038/onc.2011.496
16. Zhang ZM, Wu JF, Luo QC, et al. Pygo2 activates MDR1 expression and mediates chemoresistance in breast cancer via the Wnt/β-catenin pathway. Oncogene. 2016;35:4787–4797. doi:10.1038/onc.2016.10
17. Jang GB, Kim JY, Cho SD, et al. Blockade of Wnt/β-catenin signaling suppresses breast cancer metastasis by inhibiting CSC-like phenotype. Sci Rep. 2015;5:12465. doi:10.1038/srep12465
18. Simmons MJ, Serra R, Hermance N, et al. NOTCH1 inhibition in vivo results in mammary tumor regression and reduced mammary tumorsphere-forming activity in vitro. Breast Cancer Res. 2012;14:R126. doi:10.1186/bcr3321
19. Jurmeister S, Baumann M, Balwierz A, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. 2012;32:633–651. doi:10.1128/MCB.06212-11
20. Hu Y, Qiu Y, Yagüe E, et al. miRNA-205 targets VEGFA and FGF2 and regulates resistance to chemotherapeutics in breast cancer. Cell Death Dis. 2016;7:e2291. doi:10.1038/cddis.2016.194
21. Finlay-Schultz J, Cittelly DM, Hendricks P, et al. Progesterone downregulation of miR-141 contributes to expansion of stem-like breast cancer cells through maintenance of progesterone receptor and Stat5a. Oncogene. 2015;34:3676–3687. doi:10.1038/onc.2014.298
22. Liu R, Li J, Lai Y, et al. Hsa-miR-1 suppresses breast cancer development by down-regulating K-ras and long non-coding RNA MALAT1. Int J Biol Macromol. 2015;81:491–497. doi:10.1016/j.ijbiomac.2015.08.016
23. Kang L, Mao J, Tao Y, et al. MicroRNA-34a suppresses the breast cancer stem cell-like characteristics by downregulating Notch1 pathway. Cancer Sci. 2015;106:700–708. doi:10.1111/cas.12656
24. Li B, Lu Y, Wang H, et al. miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother. 2016;79:93–101. doi:10.1016/j.biopha.2016.01.045
25. Hui C, Yujie F, Lijia Y, et al. MicroRNA-34a and microRNA-21 play roles in the chemopreventive effects of 3,6-dihydroxyflavone on 1-methyl-1-nitrosourea-induced breast carcinogenesis. Breast Cancer Res. 2012;14:R80. doi:10.1186/bcr3194
26. Sun X, Xu C, Tang SC, et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–89. doi:10.1038/cgt.2016.3
27. Lee KH, Goan YG, Hsiao M, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315(15):2529–2538. doi:10.1016/j.yexcr.2009.06.001
28. Su JL, Chen PB, Chen YH, et al. Downregulation of microRNA miR-520h by E1A contributes to anticancer activity. Cancer Res. 2010;70(12):5096–5108. doi:10.1158/0008-5472.CAN-09-4148
29. Su CM, Wang MY, Hong CC, et al. miR-520h is crucial for DAPK2 regulation and breast cancer progression. Oncogene. 2016;35(9):1134–1342. doi:10.1038/onc.2015.168
30. Zhang M, Yang Q, Zhang L, et al. miR-302b is a potential molecular marker of esophageal squamous cell carcinoma and functions as a tumor suppressor by targeting ErbB4. J Exp Clin Cancer Res. 2014;33:10. doi:10.1186/1756-9966-33-10
31. Tian RQ1, Wang XH, Hou LJ, et al. MicroRNA-372 is down-regulated and targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human cervical cancer, which may contribute to tumorigenesis. J Biol Chem. 2011;286(29):25556–25563. doi:10.1074/jbc.M111.221564
32. Zhang W, Kong G, Zhang J, et al. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PLoS One. 2012;7(2):e31450. doi:10.1371/journal.pone.0031450
33. Zhang S, Shan C, Kong G, et al. MicroRNA-520e suppresses growth of hepatoma cells by targeting the NF-kappaB-inducing kinase (NIK). Oncogene. 2012;31(31):3607–3620. doi:10.1038/onc.2011.523
34. Wang F, Xue X, Wei J, et al. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010;103(4):567–574. doi:10.1038/sj.bjc.6605724
35. Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. doi:10.1038/nrc3458
36. Furth N, Aylon Y. The LATS1 and LATS2 tumor suppressors: beyond the Hippo pathway. Cell Death Differ. 2017;24(9):1488–1501. doi:10.1038/cdd.2017.99
37. Zhu C, Ji X, Zhang H, et al. Deubiquitylase USP9X suppresses tumorigenesis by stabilizing large tumor suppressor kinase 2 (LATS2) in the Hippo pathway. J Biol Chem. 2018;293(4):1178–1191. doi:10.1074/jbc.RA117.000392
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.