Back to Journals » OncoTargets and Therapy » Volume 9
miR-497 suppresses epithelial–mesenchymal transition and metastasis in colorectal cancer cells by targeting fos-related antigen-1
Authors Zhang N, Shen Q, Zhang P
Received 8 June 2016
Accepted for publication 29 August 2016
Published 25 October 2016 Volume 2016:9 Pages 6597—6604
DOI https://doi.org/10.2147/OTT.S114609
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Nan Zhang,1 Quan Shen,2 Pingping Zhang3
1Department of General Surgery, First Affiliated Hospital of Henan University of Chinese Medicine, 2Department of Hepatobiliary Surgery, Henan Provincial People’s Hospital, Zhengzhou, 3Department of Integrated Chinese and Western Medicine, Tianjin First Central Hospital, Tianjin, People’s Republic of China
Objective: MicroRNAs have key roles in tumor metastasis. The acquisition of metastatic capability by cancer cells is associated with epithelial–mesenchymal transition (EMT). Here, we describe the role and molecular mechanism of miR-497 in colorectal cancer (CRC) cell EMT, migration, and invasion.
Methods: Quantitative real-time polymerase chain reaction and Western blot assays were performed to detect the expression levels of miR-497 and Fos-related antigen-1 (Fra-1) in the CRC cells. HCT116 and SW480 cells with miR-497 overexpression or Fra-1 low expression were constructed by lipofection. Target prediction and luciferase reporter assays were performed to investigate whether Fra-1 is one of the targets of miR-497. Western blot and Transwell assays were performed to detect the effects of miR-497 and Fra-1 on CRC cell EMT, migration and invasion.
Results: We searched the miRanda, TargetScan, and PicTar databases and found that Fra-1, a key driver of CRC metastasis, is a potential target of miR-497. Quantitative real-time polymerase chain reaction and Western blot analysis verified downregulation of miR-497 and upregulation of Fra-1 in CRC cells. Western blot and Transwell assays showed that overexpression of miR-497 suppresses CRC cell EMT, migration, and invasion. Luciferase gene reporter assay revealed that Fra-1 is a downstream target of miR-497 as miR-497 bound directly to the 3' untranslated region of Fra-1 messenger RNA. An inverse correlation was also found between miR-497 and Fra-1 in HCT116 and SW480 cells. Furthermore, knockdown of Fra-1 recuperated the effects of miR-497 overexpression.
Conclusion: miR-497 suppresses CRC cell EMT, migration, and invasion partly by targeting Fra-1.
Keywords: colorectal cancer, miR-497, Fos-related antigen-1, epithelial–mesenchymal transition, metastasis
Introduction
Metastasis is responsible for almost 90% of cancer-associated mortality, which is a process whereby cancer cells spread from a primary site to a secondary site and form tumors.1 Metastasis involves a complex, multistep invasion–metastasis cascade.2,3 Although extensive efforts have been made, metastasis remains the most poorly understood processes in cancer biology. Recently, many studies reported that the acquisition of metastatic capability by cancer cells can be associated with epithelial–mesenchymal transition (EMT).4–6 EMT is a process whereby epithelial cells with a cobblestone phenotype acquire the characteristics of mesenchymal cells with a spindle-shaped fibroblast-like morphology. With the changes in cell phenotype, the prototypical epithelial markers E-cadherin and β-catenin decrease, and the mesenchymal markers fibronectin and vimentin increase. Upon EMT, tumor cells have the ability to invade through the basement membrane of the primary tissue and stroma, and circulate in the blood. They often become resistant to anoikis, which enables them to survive in the absence of attachment. Finally, they associate with the endothelium and extravasate to a secondary tissue and form new tumors.7,8 Although major advances in cancer therapy have been achieved, metastases remain difficult to cure due to the fact that they can be widespread, leading to tissue function damage, and they are often resistant to conventional therapy.
Colorectal cancer (CRC) is among the most common cancers and one of the most frequent causes of cancer-related deaths worldwide. In these CRC patients, it is not the primary tumor, but its metastases at distant sites are the main cause of death.9,10 Undoubtedly, a better understanding of the molecular mechanisms underlying CRC metastasis is very important to develop therapeutic strategies for metastatic CRC patients.
MicroRNAs (miRNAs) consist of about 19–22 nucleotides and post-transcriptionally modulate gene expression by base pairing to 3′ untranslated region of targeted messenger RNAs (mRNAs).11 Precise chronological and topological regulation of post-transcriptional gene silencing by miRNAs is necessary for tissue differentiation and animal development,12 and dysregulated expression is connected with various human diseases, including cancer.13 Recent emerging evidence indicates a critical role of miRNAs in cancer metastasis. miR-497, a cancer metastasis-related miRNA, has been widely reported to be dysregulated in many cancers, including CRC.14–20 Guo et al21 also reported that miR-497 inhibits CRC cell invasion by targeting insulin-like growth factor 1 receptor. Fos-related antigen-1 (Fra-1) is a member of the Fos family, which is a key driver of CRC metastasis.22–24 By target prediction analysis, Fra-1 was also found to be a potential target of miR-497. Based on these findings, we speculate that miR-497 might regulate CRC metastasis partly by targeting Fra-1. In this study, we investigated whether miR-497 targets Fra-1 to modulate EMT, invasion and migration in CRC cells.
Materials and methods
Cell culture and transfection
This study was performed with the approval of the Ethical Committee of Henan University of Chinese Medicine. The written informed consent was obtained from all patients. The normal fetal human colon epithelial cell line CRL-1831 and human CRC cell lines LoVo, RKO, HCT15, HCT28, HCT116, and SW480 were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 2 mM glutamine and 10% fetal bovine serum (HyClone, Logan, UT, USA) in a 37°C humidified atmosphere of 5% CO2.
The miR-497 mimics, small interfering RNA (siRNA) for Fra-1 (siFra-1), and their respective controls were obtained from GenePharma (Shanghai, People’s Republic of China). Cells were incubated in antibiotic-free reduced serum medium for 24 hours, and then transfected with miRNA mimics or siRNA using Lipofactamine 2000 (Invitrogen). The medium was replaced with complete medium 6 hours after transfection. Subsequent experiments were conducted 48 hours after transfection.
Quantitative real-time polymerase chain reaction
Total RNA was isolated from cultured cells using Trizol agent (Invitrogen) in accordance with the manufacturer’s instructions, and was inversely transcribed to complementary DNA by a first strand complementary DNA synthesis kit (Promega, Madison, WI, USA). miR-497 expression was detected by a mirVana miRNA detection kit (Genmed Scientifics, Arlington, MA, USA) and normalized to the level of U6 snRNA. Fra-1 mRNA level was detected by SYBR Green polymerase chain reaction (PCR) master mix on a 7500 Fast Real-time PCR system (Applied Biosystems, Foster City, CA, USA) and normalized to the level of glyceraldehyde-3-phosphate dehydrogenase.
Western blot analysis
Anti-E-cadherin, anti-β-catenin, and anti-β-actin antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-fibronectin, anti-vimentin anti-matrix metalloproteinase (MMP)-2, and anti-MMP-9 antibodies were bought from Abcam (Cambridge, MA, USA). All these antibodies were monoclonal rabbit antibodies. Briefly, cultured cells were transferred to tubes containing radioimmunoprecipitation assay buffer (Thermo Scientific, Waltham, MA, USA) and vortexed briefly. After the cells were lysed, the mixed contents were centrifuged at 14,000× g for 30 minutes at 4°C, and the lysate supernatant was collected. Proteins were denatured with loading buffer at 100°C for 3 minutes, and protein concentrations were determined using a bicinchoninic acid protein assay kit (Keygen Biotech. Co. Ltd., Nanjing, China). Western blot analysis was carried out as previously described.25
Migration and invasion assay
Migration and invasion assays were performed as described previously using transwell chambers.26
Luciferase reporter assay
A fragment of the wild-type 3′ untranslated region (3′UTR-WT) of Fra-1 containing the putative miR-497 target-sites was amplified by PCR using primers 5′-TCC GAG CTC CAC CCT AGC CAA TGT CTC CT-3′ and 5′-TGC TCT AGA GGA GCT GAA GGC TTC TCA AA-3′, and was inserted into a psiChECK-2 vector (Promega) immediately downstream to the luciferase gene sequence. Site-directed mutagenesis of the miR-497 binding sites in the Fra-1-3′UTR (3′UTR-MUT) was performed by using Quik change-mutagenesis kit (Stratagene, Heidelberg, Germany). Subsequently, psiCHECK-2 vectors including 3′UTR-WT or 3′UTR-MUT fragments of Fra-1 were transfected into miR-497-overexpressing HCT116 and SW480 cells, respectively. At 48 hours post-transfection, luciferase activity was determined by using a Dual-Luciferase Reporter Assay System (Promega).
Statistical analyses
Statistical analyses were performed using GraphPad Prism software. Values were expressed as mean ± standard error. Comparisons between the two groups were carried out by the Student’s t-test. Statistical significance was achieved at *P<0.05, **P<0.01 or ***P<0.001.
Results
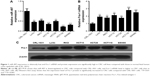
Downregulation of miR-497 and upregulation of Fra-1 exist in CRC cells
The expression levels of miR-497 and Fra-1 mRNA were detected by using quantitative real-time PCR and Fra-1 protein level was detected by using Western blot assays. MiR-497 level was decreased in CRC cells compared with that in CRL-1831 cells (Figure 1A). Fra-1 mRNA and protein levels were significantly increased in CRC cells compared with those in CRL-1831 cells (Figure 1B and C). These results indicated downregulation of miR-497 and upregulation of Fra-1 in CRC cells.
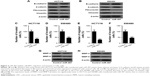
MiR-497 inhibits CRC cell EMT, migration, and invasion
To explore the effects of miR-497 on CRC cell EMT, migration, and invasion, we transfected HCT116 and SW480 cells with miR-497 mimics or negative control mimics, respectively, and then performed cell EMT, migration, and invasion detection. EMT detection showed that HCT116 and SW480 cells transfected with miR-497 mimics had obvious increases in E-cadherin and β-catenin protein expression, and marked decreases in fibronectin and vimentin protein expression compared with control cells, which indicated that miR-497 suppresses HCT116 and SW480 cell EMT (Figure 2A and B). Transwell assays revealed that miR-497 inhibits HCT116 and SW480 cell migration and invasion (Figure 2C–F). The expression levels of markers of tumor invasion, MMP-2 and MMP-9 proteins, were significantly lower in HCT116 and SW480 cells transfected with miR-497 mimics than those in control cells, which further proved that miR-497 suppresses HCT116 and cell invasion (Figure 2G and H). These data indicated that miR-497 suppresses HCT116 and SW480 cell EMT, migration, and invasion.
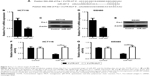
Fra-1 targeted by miR-497
To investigate the exact molecular mechanisms of miR-497 in CRC, we searched the miRanda, TargetScan, and PicTar database and found that Fra-1 is a potential target of miR-497 (Figure 3A). Then quantitative PCR, Western blot, and luciferase gene reporter assay were conducted to validate our hypothesis. As shown in Figure 3B–E, overexpression of miR-497 in HCT116 and SW480 cells caused the marked reduction in Fra-1 mRNA and protein expression. Forced expression of miR-497 inhibited luciferase activity in 3′UTR-WT, but it did not influence the luciferase activity of reporter containing 3′UTR-MUT of Fra-1 (Figure 3F and G). These data indicated that miR-497 can directly target Fra-1.
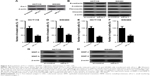
Inhibition of Fra-1 recuperated the effects of miR-497
To examine whether miR-497 regulated CRC cell EMT, migration and invasion by targeting Fra-1, we transfected HCT116 and SW480 cells with siFra-1 or si-control and then conducted cell EMT, migration, and invasion detection. Figure 4A showed that the expression levels of Fra-1 were significantly reduced in HCT116 and SW480 cells transfected with siFra-1. EMT detection showed that HCT116 and SW480 cells transfected with siFra-1 had significant increases in E-cadherin and β-catenin protein expression and remarkable decreases in fibronectin and vimentin protein expression compared with control cells (Figure 4B). Transwell assays showed that the siFra-1 groups had less invaded and migrated cells than si-control groups (Figure 4C–F). In addition, siFra-1 groups had lower levels of MMP-2 and MMP-9 proteins than si-control groups (Figure 4G and H). These results showed that Fra-1 inhibition recuperated the effects of miR-497 overexpression on CRC cell EMT, migration, and invasion, which suggested that the functional effects of miR-497 on CRC cells at least in part depends on its a direct target Fra-1.
Discussion
MiR-497 belongs to the miR-15/16/195/424/497 family, whose members share the same 3′-UTR seed sequence. As a cancer-related miRNA, miR-497 has been widely reported to act as a tumor suppressor in various tumors. For instance, miR-497 is downregulated and targets multiple cell cycle regulators to suppress the development of hepatocellular carcinoma,15 induces apoptosis of breast cancer cells,16 suppresses tumor growth and angiogenesis by targeting hepatoma-derived growth factor in non-small cell lung cancer,17 targets the insulin-like growth factor one receptor to suppress the development of human cervical cancer,18 suppresses proliferation and induces apoptosis in prostate cancer cells,19 and increases apoptosis in V-myc myelocytomatosis viral related oncogene-amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1.20
CRC is the fourth and third most common cancer in males and females, respectively, of which over 1.2 million cases are diagnosed each year worldwide with about 600,000 deaths.27 The primary cause of CRC-induced death is metastasis to the liver.10 Therefore, understanding the molecular mechanisms underlying CRC metastasis is very significant to develop novel and effective therapeutic strategies for advanced CRC patients. MiR-497 has been reported to be downregulated and inhibit CRC cell survival and invasion by targeting insulin-like growth factor 1 receptor.21,28 We further investigated whether miR-497 exerts regulatory effects on CRC metastasis by targeting other genes. We searched the miRanda, TargetScan, and PicTar databases and found that Fra-1 is also a potential target of miR-497. Fra-1, an important member of the Fos family, is frequently elevated by oncogenic signaling in various human tumors and involved in metastasis and poor prognosis. In contrast to the tumorigenic activity of c-Fos, Fra-1 seems to function in the motile and invasive phenotypes of tumor cells.29 For example, Fra-1 has been reported to promote metastasis through a variety of molecules: MMPs in breast cancer and lung epithelial cells,30,31 adenosine receptor A2b in breast cancer,32 receptor tyrosine kinase Axl in bladder cancer,33 and CD44 in mesothelioma.34 Recently, the role of Fra-1 in CRC metastasis has been widely studied. Iskit et al22 reported that Fra-1 is a key driver of CRC metastasis and a Fra-1 classifier predicts disease-free survival. Diesch et al24 found that Fra-1 is strongly expressed in tumor cells at the invasive front of human CRC, and that its depletion suppresses mesenchymal-like features in CRC cells in vitro. Liu et al23 found that aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes CRC aggressiveness through EMT. Based on these findings, we speculated that miR-497 might regulate CRC cell EMT, migration, and invasion in part by targeting Fra-1. In this study, we detected the expression levels of miR-497 and Fra-1 in CRC cell lines, and found downregulation of miR-497 and upregulation of Fra-1 in CRC cells, which were completely consistent with previous reports.21–24,28 We further investigated the effects of miR-497 on CRC cell EMT, migration, and invasion, and found that overexpression of miR-497 inhibits CRC cell EMT, migration, and invasion. We performed luciferase assay and confirmed that Fra-1 can be directly targeted by miR-497. The expression levels of Fra-1 mRNA and protein were both regulated by miR-497, and silencing Fra-1 by siRNA recapitulated the effects of miR-497 on CRC cells. These results confirmed our speculation that miR-497 inhibits CRC cell EMT, migration, and invasion partly by targeting Fra-1.
All together, this study shows that miR-497 is downregulated in CRC cells and inversely related to cell EMT, migration, and invasion, while Fra-1, a potential oncogene in CRC, is upregulated and positively correlated with CRC cell EMT, migration, and invasion. The role of miR-497 is partially mediated by the target gene Fra-1. These findings facilitate a better understanding of the molecular mechanisms underlying CRC metastasis and provide novel potential therapeutic targets for metastatic CRC.
Acknowledgments
This work was supported by grants from the Science and Technology Project of Henan Province (No 112102310700) and the Natural Science Foundation of Education Department of Henan Province (No 2011A320007).
Disclosure
The authors report no conflicts of interest in this work.
References
Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331(6024):1559–1564. | ||
Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. | ||
Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12(8):895–904. | ||
Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. | ||
Thompson EW, Newgreen DF. Carcinoma invasion and metastasis: a role for epithelial–mesenchymal transition? Cancer Res. 2005;65(14):5991–5995. | ||
Christiansen JJ, Rajasekaran AK. Reassessing epithelial to mesenchymal transition as a prerequisite for carcinoma invasion and metastasis. Cancer Res. 2006;66(17):8319–8326. | ||
Geiger TR, Peeper DS. Metastasis mechanisms. BBA-Rev Cancer. 2009;1796(2):293–308. | ||
Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. | ||
Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1688–1694. | ||
Welch JP, Donaldson G. The clinical correlation of an autopsy study of recurrent colorectal cancer. Ann Surg. 1979;189(4):496–502. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132(21):4653–4662. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Tokarz P, Blasiak J. The role of microRNA in metastatic colorectal cancer and its significance in cancer prognosis and treatment. Acta Biochim Pol. 2012;59(4):467–474. | ||
Furuta M, Kozaki K-I, Tanimoto K, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8(3):e60155. | ||
Shen L, Li J, Xu L, et al. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3(3):475–480. | ||
Zhao WY, Wang Y, An ZJ, et al. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem Biophys Res Commun. 2013;435(3):466–471. | ||
Luo M, Shen D, Zhou X, Chen X, Wang W. MicroRNA-497 is a potential prognostic marker in human cervical cancer and functions as a tumor suppressor by targeting the insulin-like growth factor 1 receptor. Surgery. 2013;153(6):836–847. | ||
Wang L, Li B, Li L, Wang T. MicroRNA-497 suppresses proliferation and induces apoptosis in prostate cancer cells. Asian Pac J Cancer Prev. 2013;14(6):3499–3502. | ||
Creevey L, Ryan J, Harvey H, et al. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol Cancer. 2013;12(1):23. | ||
Guo S, Jiang C, Wang G, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32(15):1910–1920. | ||
Iskit S, Schlicker A, Wessels L, Peeper DS. Fra-1 is a key driver of colon cancer metastasis and a Fra-1 classifier predicts disease-free survival. Oncotarget. 2015;6(41):43146–43161. | ||
Liu H, Ren G, Wang T, et al. Aberrantly expressed Fra-1 by IL-6/STAT3 transactivation promotes colorectal cancer aggressiveness through epithelial–mesenchymal transition. Carcinogenesis. 2015;36(4):459–468. | ||
Diesch J, Sanij E, Gilan O, et al. Widespread FRA1-dependent control of mesenchymal transdifferentiation programs in colorectal cancer cells. PLoS One. 2014;9(3):e88950. | ||
Tian Y, Luo A, Cai Y, et al. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J Biol Chem. 2010;285(11):7986–7994. | ||
Yue D, Zhang Z, Li J, et al. Transforming growth factor-beta1 promotes the migration and invasion of sphere-forming stem-like cell subpopulations in esophageal cancer. Exp Cell Res. 2015;336(1):141–149. | ||
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. | ||
Corté H, Manceau G, Blons H, Laurent-Puig P. MicroRNA and colorectal cancer. Digest Liver Dis. 2012;44(3):195–200. | ||
Young MR, Colburn NH. Fra-1 a target for cancer prevention or intervention. Gene. 2006;379:1–11. | ||
Belguise K, Kersual N, Galtier F, Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene. 2005;24(8):1434–1444. | ||
Adiseshaiah P, Vaz M, Machireddy N, Kalvakolanu DV, Reddy SP. A Fra-1-dependent, matrix metalloproteinase driven EGFR activation promotes human lung epithelial cell motility and invasion. J Cellular Physiol. 2008;216(2):405–412. | ||
Desmet CJ, Gallenne T, Prieur A, et al. Identification of a pharmacologically tractable Fra-1/ADORA2B axis promoting breast cancer metastasis. Proc Natl Acad Sci U S A. 2013;110(13):5139–5144. | ||
Sayan AE, Stanford R, Vickery R, et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene. 2012;31(12):1493–1503. | ||
Ramos-Nino ME, Blumen SR, Pass H, Mossman BT. Fra-1 governs cell migration via modulation of CD44 expression in human mesotheliomas. Mol Cancer. 2007;6(1):81. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.