Back to Journals » OncoTargets and Therapy » Volume 12
miR-492 Promotes Cancer Progression by Targeting GJB4 and Is a Novel Biomarker for Bladder Cancer
Authors Wang K, Lü H, Qu H, Xie Q, Sun T, Gan O, Hu B
Received 16 July 2019
Accepted for publication 7 December 2019
Published 24 December 2019 Volume 2019:12 Pages 11453—11464
DOI https://doi.org/10.2147/OTT.S223448
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Federico Perche
Kai Wang, Hang Lü, Hongchen Qu, Qingpeng Xie, Tao Sun, Ou Gan, Bin Hu
Department of Urology Surgery, Liaoning Province Cancer Hospital and Institute (Cancer Hospital of China Medical University), Shenyang 110004, People’s Republic of China
Correspondence: Bin Hu
Department of Urology Surgery, Liaoning Province Cancer Hospital and Institute (Cancer Hospital of China Medical University), 44 Xiaoheyan Street, Shen-Yang 110004, People’s Republic of China
Email [email protected]
Background: Bladder cancer is the most common urinary system malignancy in the United States and is characterized by its diverse prognosis and high recurrence rate. However, the molecular mechanisms underlying its progression remain unknown. Accumulating evidence suggests a critical role for miRNAs in bladder cancer progression.
Methods and results: In this study, we found that miR-492 expression levels were significantly higher in bladder cancer tissue and the serum of bladder cancer patients by bioinformatics analysis and a panel of clinical samples. The results of receiver operating characteristic curve analysis suggested the potential diagnostic value of serum miR-492 for bladder cancer. In vitro and in vivo functional assays showed that knockdown of miR-492 suppressed proliferation and metastasis of bladder cancer cells. Gap junction beta-4 protein was predicted to be a direct target of miR-492, which was validated using a luciferase reporter assay. Further cellular functional assays showed that suppression of miR-492 abrogated bladder cancer cell proliferation and metastasis by targeting gap junction beta-4 protein.
Conclusion: miR-492 promotes cancer progression by targeting GJB4 and is a novel biomarker for bladder cancer.
Keywords: miRNAs, miR-492, bladder cancer, GJB4, biomarker
Introduction
Bladder cancer is the most common urinary system malignancy in the United States.1 Estimated new cases of bladder cancer in male patients are three times more than that of female patients.2 Clinical outcomes for bladder cancer differ significantly with different clinical and prognostic variables. The five-year relative survival rates for bladder cancer are 96% for carcinoma in situ, 70% for localized, 35% for regional, and 5% for distant.2 Unfortunately, 40 to 80% of patients with carcinoma in situ will have disease recurrence or progression after standard clinical treatment.3 Elevation of the overall survival rate of bladder cancer patients requires further elucidation of the molecular mechanisms underlying disease progression and identification of novel diagnostic biomarkers.4
MicroRNAs (miRNAs) are small noncoding RNAs that can mediate the degradation of their target messenger RNAs (mRNAs) or inhibit their translation by binding to seed sequences in their 3ʹ-untranslated regions.5–7 Multiple studies have reported that aberrant expression of miRNAs correlates closely with disease progression in multiple cancer types.8–13 Furthermore, miRNAs are also enriched in human body fluids (e.g., blood, saliva, bile).14–17 The expression profile of miRNAs in cancer patient blood could reflect the pathological status, which makes miRNAs promising candidates for novel biomarkers in cancer.18–20 miR-492 has been reported to promote cancer progression in several cancer types.21–27 However, the role of this miRNA in bladder cancer and its potential as a biomarker has not been reported.
In this study, we found that miR-492 was significantly upregulated in bladder cancer tissue and serum from bladder cancer patients using bioinformatics analysis of two different Gene Expression Omnibus (GEO) miRNA microarray datasets. These findings were validated with a panel of clinical samples, suggesting that miR-492 expression is significantly higher in bladder cancer tissue and patient serum than paracancerous tissues and serum from healthy participants. Receiver operating characteristic (ROC) curve analysis showed the potential diagnostic value of serum miR-492 for bladder cancer. In vitro and in vivo experiments demonstrated that knockdown of miR-492 could suppress proliferation, migration, invasion, and the epithelial-mesenchymal transition, and induce apoptosis in bladder cancer cells by targeting gap junction beta-4 protein (GJB4).
Materials and Methods
Ethics Approval and Consent to Participate
Animal studies and usage of clinical samples were approved by the ethics committee of the Liaoning Cancer Hospital and Institute (Affiliated Cancer Hospital of China Medical University). All participants received and signed a detailed written informed consent form. Animal handling procedures were performed according to the standards of the local ethics committee.
Bioinformatics Analysis
Two publicly available miRNA microarray datasets (GSE40355, 16 bladder cancer tissue samples and eight normal bladder tissue samples; GSE112264, 50 bladder cancer serum samples and 40 normal control serum samples) were downloaded from the public Gene Expression Omnibus database. Differentially expressed miRNAs were analyzed with the R language package edgeR. Heatmaps of the differentially expressed miRNAs were visualized using the R language package pheatmap. The miR-492 expression levels in the two datasets were visualized using GraphPad Prism 7.
Cell Lines and Cell Culture
The human bladder cancer cell lines (UM-UC-3 and T24) were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (SIBCB). Cells were cultured in RPMI-1640 (Gibco, NY, USA) supplemented with 10% fetal bovine serum (Gibco).
Reverse Transcription Quantitative Polymerase Chain Reaction
Total RNA from cell lines and ground tissues was isolated with TRIzol reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. Serum miRNA was isolated with the Serum (Plasma) miRNA Isolation Kit (Foregene, Chengdu, China) according to the manufacturer’s protocol. The concentration and quality of the isolated RNA were determined using the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). For miR-492 detection, total RNA was converted into cDNA using the miRNA 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). For GJB4 detection, cDNA was generated from total RNA using the RT SuperMix for qPCR kit (Vazyme). miR-492 and GJB4 expression were measured by qPCR using the SYBR qPCR Master Mix kit (Vazyme). The primer sequences were: Hsa-miR-492 forward, 5ʹ-GGGGTACCCCCTGGCTGGAACAGAAGAT-3ʹ; Hsa-miR-492 reverse, 5ʹ-CCCAAGCTTCCCTGGTCTTGGCTGGGATC-3ʹ; U6 forward, 5ʹ-CGCTTCGGCAGCACATATAC-3ʹ; U6 reverse, 5ʹ-TTCACGAATTTGCGTGTCATC-3ʹ; GJB4 forward, 5ʹ-TCAATCGCACCAGCATTAAG-3ʹ; GJB4 reverse, 5ʹ-GGGGGA CCTGTTGATCTTATC-3ʹ; GAPDH forward, 5ʹ-ACAACTTTGGTATCGTGGAAGG-3ʹ; GAPDH reverse, 5ʹ-GCCATCACGCCACAGTTTC-3ʹ.
Western Blotting Assay
Cells and ground tissue were resuspended in RIPA buffer supplemented with protease inhibitor for 20 min on ice. Protein concentration was determined using the bicinchoninic acid protein assay kit (Beyotime, Tianjin, China) according to the manufacturer’s instructions. Total protein (40 μg) from each sample was separated on a polyacrylamide slab gel and transferred to a PVDF membrane (Bio-Rad, CA, USA). Membranes were incubated overnight at 4°C with primary antibody against GJB4 (1:1000, Abcam, Cambridge, UK), β-actin (1:8000, ProteinTech, Rosemont, USA). Blots were incubated with horseradish peroxidase-conjugated secondary antibody (1:2000, ProteinTech). Protein bands were visualized using the enhanced BeyoECL Plus kit (Beyotime).
Transfection Experiments
Synthetic miR-492 mimics, miRNA mimic negative control, miR-492 inhibitor (shRNA plasmids), negative control RNA inhibitor (shRNA plasmids), GJB4 siRNA, and negative control siRNA were purchased from Genepharma Biotechnology (Genepharma, Shanghai, China). Transfection experiments were performed using Lipofectamine 3000 reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Cell Proliferation Assay
The cell proliferation assay was performed using the Cell Counting Kit-8 (CCK-8, Dojindo, Tokyo, Japan) and Cell-Light 5-ethynyl-2ʹ-deoxyuridine (EdU, Beyotime) DNA cell proliferation assays according to the manufacturers’ protocols. For the EdU assay, nuclei were stained with Hoechst 33342, and the cells were visualized under a fluorescence microscope. Exposure times were 30 sec for visualization of Apollo and 0.180 sec for nuclei.
Flow Cytometric Apoptosis Analysis
For apoptosis analysis, cells were collected after experimental treatment and washed with PBS. The cells were then stained with the FITC/Annexin V Apoptosis Detection Kit I (BD Biosciences, NY, USA) and analyzed by flow cytometry (Thermo Fisher Scientific).
In vitro Migration Assay
For wound healing migration assays, cells with corresponding experimental treatment were cultured in 6-well plates until confluent. A wound was made across the cell monolayer using a pipette tip. The wounded cell monolayer was then cultured in 2.5% serum medium. Photos of the wound were taken at 0 and 18 h using light microscopy.
In vitro Invasion Assay
For invasion assays using Transwells (24-well, 8-μm pore size filters, Costar, NY, USA), 2×104 cells with corresponding experimental treatment in serum- and antibiotic-free medium were added to the upper chambers pre-coated with Matrigel (dilution 1:8, BD Biosciences). The lower chambers were filled with RPMI-1640 containing 10% serum and antibiotics. After co-culture for the indicated times, cells and Matrigel remaining in the upper surface were removed, and cells that invaded the lower surface were fixed with 4% paraformaldehyde. Fixed cells were stained with crystal violet and counted using light microscopy.
Luciferase Reporter Assay
Putative miR-492 targets were predicted using TargetScan (http://targetscan.org/). Based on this analysis, GJB4 was predicted as a potential miR-492 target. The 3ʹ-UTR of GJB4 containing the wild-type or mutant miR-492 binding site was constructed by Genechem Biotechnology (Shanghai, China). A total of 3×104 UM-UC-3 or T24 cells were seeded in triplicate in 24-well plates and co-transfected with the corresponding plasmids and miR-492 mimics. Luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA) according to the manufacturer’s protocol.
Bladder Cancer Xenografts
Subcutaneous tumors were generated in 5-week-old female BALB/c nude mice (Huafukang, Beijing, China). UM-UC-3 cells (8×106) transfected with miR-492 inhibitor or negative control RNA inhibitor were subcutaneously injected in 150 μL PBS into the right axilla area. Tumors were measured once per week using a Vernier caliper. Mice were euthanized four weeks after the inoculation, and the tumors excised and measured. Tumor volume was calculated using the following equation: (L×W2)/2 where L was the length of the long axis, and W was the length of the short axis.
Statistical Analysis
The Student’s t-test was performed using SPSS Statistics 19.0. P-values of <0.05 were considered statistically significant. The receiver operating characteristic curve analysis was performed using GraphPad Prism 7. Statistical charts were generated with GraphPad Prism 7.
Results
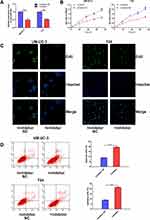
miR-492 Expression Is Significantly Upregulated in Bladder Cancer Tissue and Bladder Cancer Patient Serum
We analyzed the data from two independent publicly available GEO microarray datasets (Figure 1A and B). We found that miR-492 was significantly upregulated in bladder cancer tissue and bladder cancer patient serum (Figure 1C and D). We validated these results using a panel of clinical samples (Table 1). Analysis of this panel showed that miR-492 expression levels were higher in bladder cancer tissue and bladder cancer patient serum than in paracancerous tissue or serum from healthy participants (Figure 1E and F). The results of the ROC analysis showed the potential diagnostic value of serum miR-492 levels in bladder cancer (Figure 1G).
 |
Table 1 Clinical Characteristics of 23 Bladder Cancer Patients and Healthy Participants in This Study |
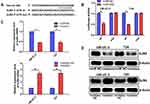
Knockdown of miR-492 Suppresses Proliferation and Induces Apoptosis of Bladder Cancer Cells
Based on expression analysis, we investigated the role of miR-492 in bladder cancer progression by suppressing miR-492 expression in UM-UC-3 and T24 cells by transfection of miR-492 inhibitors (Figure 2A). Knockdown of miR-492 expression suppressed proliferation (Figure 2B and C) and induced apoptosis (Figure 2D) of UM-UC-3 and T24 cells. Furthermore, knockdown of miR-492 expression abrogated the growth of UM-UC-3 xenografts (Figure 3A–D).
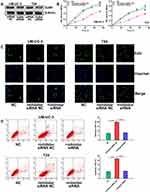
Knockdown of miR-492 Suppresses Bladder Cancer Cell Metastasis in vitro
We investigated the effect of miR-492 on the metastasis of bladder cancer cells. In vitro migration and invasion assays showed that knockdown of miR-492 suppressed migration and invasion of bladder cancer cells (Figure 4A and B).
GJB4 Is a Direct Target Gene of miR-492 in Bladder Cancer Cells
We predicted potential target genes of miR-492 by bioinformatics analysis and selected GJB4 for further analysis (Figure 5A). Luciferase activity from a reporter plasmid carrying wild-type, but not mutant, GJB4 3ʹ-UTR was significantly decreased by miR-492 overexpression in UM-UC-3 and T24 cells (Figure 5B). Furthermore, miR-492 overexpression reduced GJB4 expression, and miR-492 knockdown increased GJB4 expression in both cell lines at both the mRNA and protein level (Figure 5C and D). These results suggest that GJB4 is a direct target gene for miR-492 in bladder cancer cells.
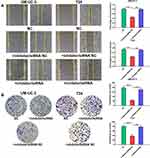
miR-492 Suppresses Proliferation and Metastasis of Bladder Cancer Cells by Targeting GJB4
To determine whether the functions of miR-492 are achieved by its targeting of GJB4, we decreased miR-492 and GJB4 expression by transfection of an miR-492 inhibitor and GJB4 siRNA, respectively, or corresponding controls (Figure 6A). In vitro functional experiments showed that the inhibitor/siRNA group had a similar phenotype to that of the control group. In contrast, the inhibitor/NC siRNA group displayed decreased proliferation, migration, invasion, and increased apoptosis (Figures 6B–D and 7A, B). These results suggest that miR-492 promoted proliferation and metastasis of bladder cancer cells by targeting GJB4.
Discussion
Though many countries, such as the United States, have developed standard screening, early detection, and clinical therapy for patients with bladder cancer, the prognosis of these patients differs significantly. For patients with local invasion and distant metastasis at the time of diagnosis, surgery provides limited efficacy. Furthermore, bladder cancer is characterized by a high recurrence rate after surgery. Even for patients with carcinoma in situ, 40 to 80% will have disease recurrence or progression after standard clinical treatment. Therefore, to increase the overall survival rates for bladder cancer patients, further elucidation of the underlying molecular mechanisms is required.
Several studies have reported the tumor-promoting effect of miR-492 in other cancer types. miR-492 promoted prostate cancer growth by repressing SOCS2, hepatoblastoma metastasis via CD44, and breast cancer proliferation by suppressing SOX7.21,24 Aberrant miR-492 expression has also been correlated with chemoresistance in cervical and colon cancer.26,28 In contrast, circular RNA circ0072309 inhibited proliferation and invasion of breast cancer cells by targeting miR-492.22 These studies demonstrated the tumor-promoting activity of miR-492. However, the role of miR-492 in bladder cancer was not reported.
In this study, we found that miR-492 was upregulated in both bladder cancer tissue and bladder cancer patient serum by bioinformatics analysis, which was validated in a panel of clinical samples. Considering that miR-492 was enriched in bladder cancer patient serum, we further studied its potential as a diagnostic biomarker for this disease. The results from the ROC analysis showed the potential diagnostic value of serum miR-492 for bladder cancer detection. The results of the functional analysis showed the significant tumor-promoting activity of miR-492 as miR-492 knockdown suppressed proliferation and metastasis of bladder cancer cells. These data suggest that miR-492 may act as a broad-spectrum tumor promoter and serve as a novel target for cancer therapy. The results of this study validated GJB4 as a direct target of miR-492. Further functional analysis showed that miR-492 suppressed proliferation and metastasis of bladder cancer cells by targeting GJB4. Multiple studies have reported a close correlation between dysregulated gap junction protein family expression and cancer progression.29–33 However, only one study has reported a role for GJB4 in lung cancer progression.34 Our results may provide some useful data for future studies on the GJB4.
In conclusion, our study was the first to report the aberrant expression and tumor-promoting activity of miR-492 via GJB4 in bladder cancer. Our results may provide evidence for further development of miR-492 as a novel biomarker and therapeutic target for bladder cancer.
Acknowledgment
The study was funded by National Fund of China Foreign Experts Bureau (20172100003).
Author Contributions
KW performed most of the experiment and wrote the paper. HL, HQ, QX, TS, and OG collected clinical samples and performed some of the experiments. BH designed the study, analyzed the data and revised the paper. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.v69.1
3. Barton MK. High morbidity and mortality found for high-risk, non-muscle-invasive bladder cancer. CA Cancer J Clin. 2013;63(6):371–372. doi:10.3322/caac.21201
4. Wang B, Pan W, Yang M, et al. Programmed death ligand-1 is associated with tumor infiltrating lymphocytes and poorer survival in urothelial cell carcinoma of the bladder. Cancer Sci. 2019;110(2):489–498. doi:10.1111/cas.2019.110.issue-2
5. Herzog K, Bandiera S, Pernot S, et al. Functional microRNA screen uncovers O-linked N-acetylglucosamine transferase as a host factor modulating hepatitis C virus morphogenesis and infectivity. Gut. 2019. doi:10.1136/gutjnl-2018-317423.
6. Zhang J, Bai R, Li M, et al. Excessive miR-25-3p maturation via N(6)-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat Commun. 2019;10(1):1858. doi:10.1038/s41467-019-09712-x
7. Bian H, Zhou Y, Zhou D, Zhang Y, Shang D, Qi J. The latest progress on miR-374 and its functional implications in physiological and pathological processes. J Cell Mol Med. 2019;23(5):3063–3076. doi:10.1111/jcmm.2019.23.issue-5
8. Wang J, Wen T, Li Z, et al. MicroRNA-1224 inhibits tumor metastasis in intestinal-type gastric cancer by directly targeting FAK. Front Oncol. 2019;9:222. doi:10.3389/fonc.2019.00222
9. Takahashi RU, Prieto-Vila M, Kohama I, Ochiya T. Development of miRNA-based therapeutic approaches for cancer patients. Cancer Sci. 2019;110(4):1140–1147. doi:10.1111/cas.13965
10. Cao L, Jin H, Zheng Y, et al. DANCR-mediated microRNA-665 regulates proliferation and metastasis of cervical cancer through the ERK/SMAD pathway. Cancer Sci. 2019;110(3):913–925. doi:10.1111/cas.13921
11. Hu J, Zhao W, Huang Y, Wang Z, Jiang T, Wang L. MiR-1180 from bone marrow MSCs promotes cell proliferation and glycolysis in ovarian cancer cells via SFRP1/Wnt pathway. Cancer Cell Int. 2019;19:66. doi:10.1186/s12935-019-0751-z
12. Lu YF, Yu JR, Yang Z, et al. Promoter hypomethylation mediated upregulation of MicroRNA-10b-3p targets FOXO3 to promote the progression of esophageal squamous cell carcinoma (ESCC). J Exp Clin Cancer Res. 2018;37(1):301. doi:10.1186/s13046-018-0966-1
13. Ramayanti O, Verkuijlen S, Novianti P, et al. Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections. Int J Cancer. 2019;144(10):2555–2566. doi:10.1002/ijc.v144.10
14. Lu Z, He Q, Liang J, et al. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol Ther Nucleic Acids. 2019;16:471–480. doi:10.1016/j.omtn.2019.03.012
15. Bahmanpour Z, Sheervalilou R, Choupani J, Shekari Khaniani M, Montazeri V, Mansoori Derakhshan S. A new insight on serum microRNA expression as novel biomarkers in breast cancer patients. J Cell Physiol. 2019. doi:10.1002/jcp.28656
16. Bhagirath D, Yang TL, Tabatabai ZL, et al. Role of a novel race-related tumor suppressor microRNA located in frequently deleted chromosomal locus 8p21 in prostate cancer progression. Carcinogenesis. 2019;40:633–642. doi:10.1093/carcin/bgz058
17. Wang J, Raimondo M, Guha S, et al. Circulating microRNAs in pancreatic juice as candidate biomarkers of pancreatic cancer. J Cancer. 2014;5(8):696–705. doi:10.7150/jca.10094
18. Usuba W, Urabe F, Yamamoto Y, et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019;110(1):408–419. doi:10.1111/cas.13856
19. Liu H, Li PW, Yang WQ, et al. Identification of non-invasive biomarkers for chronic atrophic gastritis from serum exosomal microRNAs. BMC Cancer. 2019;19(1):129. doi:10.1186/s12885-019-5328-7
20. Shiino S, Matsuzaki J, Shimomura A, et al. Serum miRNA-based prediction of axillary lymph node metastasis in breast cancer. Clin Cancer Res. 2019;25(6):1817–1827. doi:10.1158/1078-0432.CCR-18-1414
21. Shi LP, Liang M, Li FF, et al. MiR-492 exerts tumor-promoting function in prostate cancer through repressing SOCS2 expression. Eur Rev Med Pharmacol Sci. 2019;23(3):992–1001. doi:10.26355/eurrev_201902_16986
22. Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–1041. doi:10.2147/CMAR.S186857
23. Ding HX, Lv Z, Yuan Y, Xu Q. MiRNA polymorphisms and cancer prognosis: a systematic review and meta-analysis. Front Oncol. 2018;8:596. doi:10.3389/fonc.2018.00596
24. von Frowein J, Hauck SM, Kappler R, et al. MiR-492 regulates metastatic properties of hepatoblastoma via CD44. Liver Int. 2018;38(7):1280–1291. doi:10.1111/liv.2018.38.issue-7
25. Liu M, An J, Huang M, et al. MicroRNA-492 overexpression involves in cell proliferation, migration, and radiotherapy response of cervical squamous cell carcinomas. Mol Carcinog. 2018;57(1):32–43. doi:10.1002/mc.v57.1
26. Peng L, Zhu H, Wang J, et al. MiR-492 is functionally involved in Oxaliplatin resistance in colon cancer cells LS174T via its regulating the expression of CD147. Mol Cell Biochem. 2015;405(1–2):73–79. doi:10.1007/s11010-015-2397-z
27. Jiang J, Zhang Y, Yu C, Li Z, Pan Y, Sun C. MicroRNA-492 expression promotes the progression of hepatic cancer by targeting PTEN. Cancer Cell Int. 2014;14(1):95. doi:10.1186/s12935-014-0095-7
28. Liu M, Wang Z, Liu Q, Zhu H, Xu N. Expression of Micro-RNA-492 (MiR-492) in human cervical cancer cell lines is upregulated by transfection with wild-type P53, irradiation, and 5-fluorouracil treatment in vitro. Med Sci Monit. 2018;24:7750–7758. doi:10.12659/MSM.911585
29. Xiao H, Liu W, Zhao Z, Zhang Y, Song Y, Luo B. Single nucleotide polymorphism rs2274084 of gap junction protein beta 2 gene among Epstein-Barr virus-associated tumors. Cancer Biomark. 2018;21(3):499–504. doi:10.3233/CBM-170078
30. Kou Y, Ji L, Wang H, et al. Connexin 43 upregulation by dioscin inhibits melanoma progression via suppressing malignancy and inducing M1 polarization. Int J Cancer. 2017;141(8):1690–1703. doi:10.1002/ijc.v141.8
31. Phillips SL, Williams CB, Zambrano JN, Williams CJ, Yeh ES. Connexin 43 in the development and progression of breast cancer: what’s the connection? (Review). Int J Oncol. 2017;51(4):1005–1013. doi:10.3892/ijo.2017.4114
32. Ai XL, Chi Q, Qiu Y, et al. Gap junction protein connexin43 deregulation contributes to bladder carcinogenesis via targeting MAPK pathway. Mol Cell Biochem. 2017;428(1–2):109–118. doi:10.1007/s11010-016-2921-9
33. Ruch RJ. Connexin43 suppresses lung cancer stem cells. Cancers (Basel). 2019;11(2):175. doi:10.3390/cancers11020175
34. Lin YP, Wu JI, Tseng CW, Chen HJ, Wang LH. Gjb4 serves as a novel biomarker for lung cancer and promotes metastasis and chemoresistance via Src activation. Oncogene. 2019;38(6):822–837. doi:10.1038/s41388-018-0471-1
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.