Back to Journals » OncoTargets and Therapy » Volume 12
miR-424-5p Promotes Proliferation, Migration and Invasion of Laryngeal Squamous Cell Carcinoma
Authors Li Y , Liu J, Hu W, Zhang Y, Sang J, Li H , Ma T, Bo Y, Bai T, Guo H, Lu Y, Xue X, Niu M , Ge S, Wen S, Wang B , Gao W , Wu Y
Received 23 July 2019
Accepted for publication 18 November 2019
Published 29 November 2019 Volume 2019:12 Pages 10441—10453
DOI https://doi.org/10.2147/OTT.S224325
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Takuya Aoki
Yujun Li,1–4,* Jie Liu,5,* Wanglai Hu,6,* Yuliang Zhang,1–4,* Jiangwei Sang,1–4 Huizheng Li,7 Teng Ma,8 Yunfeng Bo,9 Tao Bai,10 Huina Guo,1–4 Yan Lu,11 Xuting Xue,1–4 Min Niu,1–4 Shanshan Ge,12 Shuxin Wen,1–4 Binquan Wang,1–4 Wei Gao,1–4 Yongyan Wu1–4
1Shanxi Key Laboratory of Otorhinolaryngology Head and Neck Cancer, Shanxi Medical University, Taiyuan, Shanxi 030001, People’s Republic of China; 2Department of Otolaryngology Head & Neck Surgery, The First Hospital of Shanxi Medical University, Taiyuan, Shanxi 030001, People’s Republic of China; 3Otolaryngology Head & Neck Surgery Research Institute, Shanxi Medical University, Taiyuan, Shanxi 030001, People’s Republic of China; 4The Key Scientific and Technological Innovation Platform for Precision Diagnosis and Treatment of Head and Neck Cancer, Shanxi Province, Taiyuan 030001, Shanxi, People’s Republic of China; 5Department of Head and Neck Surgical Oncology, National Cancer Center, National Clinical Research Center for Cancer, Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing 100021, People’s Republic of China; 6School of Basic Medical Science, Anhui Medical University, Hefei, Anhui 230027, People’s Republic of China; 7Department of Otolaryngology Head & Neck Surgery, Dalian Municipal Friendship Hospital, Dalian, Liaoning 116001, People’s Republic of China; 8Department of Cellular and Molecular Biology, Beijing Chest Hospital, Beijing Tuberculosis and Thoracic Tumor Research Institute, Capital Medical University, Beijing 101149, People’s Republic of China; 9Department of Pathology, Shanxi Cancer Hospital, Shanxi Medical University, Taiyuan, Shanxi 030000, People’s Republic of China; 10Department of Pathology, The First Hospital of Shanxi Medical University, Taiyuan, Shanxi 030001, People’s Republic of China; 11Department of Otolaryngology Head & Neck Surgery, The First Hospital, Jinzhou Medical University, Jinzhou 121001, Liaoning, People’s Republic of China; 12Health Management Center, the First Hospital of Shanxi Medical University, Taiyuan 030001, Shanxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yongyan Wu; Wei Gao
Shanxi Key Laboratory of Otorhinolaryngology Head and Neck Cancer, The First Hospital of Shanxi Medical University, No. 85, South Jiefang Road, Taiyuan, Shanxi 030001, People’s Republic of China
Tel +86 351-4867076
Fax +86 351-4639218
; +86 351-4639218
Email [email protected]; [email protected]
Background: Recent studies revealed that miR-424-5p regulates the malignant behavior of multiple cancer types. However, the expression and function of miR-424-5p in laryngeal squamous cell carcinoma (LSCC) is unclear.
Purpose: This study aimed to evaluate the association of miR-424-5p level with clinical features of LSCC and investigate the effect and potential mechanism of miR-424-5p on LSCC progression.
Methods: The expression of miR-424-5p in LSCC and paired adjacent normal margin (ANM) tissues from 106 patients with LSCC were analyzed by quantitative PCR (qPCR), and clinical significance was analyzed. Target genes of miR-424-5p were predicted, followed by functional annotation. The functional role of miR-424-5p in LSCC was investigated by molecular and cellular experiments with LSCC cell lines, with flow cytometry used for cell cycle analysis. In addition, miR-424-5p regulation of the predicted target gene cell adhesion molecule 1 (CADM1) was validated by qPCR, Western blot analysis and luciferase reporter assay.
Results: miR-424-5p was upregulated in LSCC versus ANM tissues. High miR-424-5p level was significantly associated with poor differentiation, advanced tumor stage and cervical lymph node metastasis. Bioinformatics analysis showed that miR-424-5p target genes are mainly enriched in biological processes of the cell cycle, cell division, and negative regulation of cell migration, and were involved in multiple cancer-related pathways. Overexpression of miR-424-5p promoted proliferation, migration, invasion, and adhesion of LSCC cells and affected the cell cycle progression. Additionally, CADM1 was a direct target of miR-424-5p in LSCC cells.
Conclusion: miR-424-5p functions as an oncogene to promote the aggressive progression of LSCC, and CADM1 is a direct downstream target of miR-424-5p in LSCC cells. miR-424-5p may be a potential therapeutic target in LSCC.
Keywords: laryngeal squamous cell carcinoma, miR-424-5p, proliferation, migration and invasion, oncogenic miRNA
Introduction
Laryngeal squamous cell carcinoma (LSCC) represents the second-highest incidence of head and neck squamous cell carcinoma.1,2 The etiological factors mainly include tobacco smoking, alcohol consumption, air contamination, chronic inflammation, and human papillomavirus infection.3 The general mechanism of LSCC involves an imbalance in oncogenes and tumor suppressor genes and disordered immune mechanisms.4,5 However, the detailed and specific pathogenesis of LSCC is not clear.
Surgical removal of tumors combined with radiotherapy and chemotherapy is the usual treatment for LSCC. Despite improvements in diagnostic and therapeutic strategies, there has been no significant improvement in LSCC survival over the past 30 years.6 The current challenge demands the discovery of accurate and non-invasive biomarkers for diagnosis, prognosis and prediction of recurrence. This is needed to improve the clinical management of LSCC and identify new molecular targets for clinical therapy.
MicroRNAs (miRNAs) are small noncoding RNA molecules that are important in many cellular processes, including development, differentiation, cell cycle regulation, and apoptosis.7,8 miRNAs may act as oncogenes or tumor suppressor genes to regulate tumor tumorigenesis and progression. Microarray and RNA-sequencing revealed a number of differentially expressed miRNAs in LSCC.9,10 Furthermore, recent studies investigated the role of miRNAs in regulating LSCC. miR-145-5p was downregulated in LSCC, and in vitro and in vivo experiments showed that its overexpression inhibited the proliferation, migration and invasion of LSCC cells.11 miR-204 suppressed the invasion and metastasis of LSCC.12 However, miR-301-3p and miR-132 enhanced the cell proliferation and tumor growth of LSCC.13,14
Downregulation of miR-424-5p has been reported in cervical cancer, epithelial ovarian cancer, and liver cancer. miR-424-5p functions as a tumor suppressor in these cancers.15–17 However, miR-424-5p was found significantly upregulated in pancreatic cancer and increased the proliferation, migration, and invasion of pancreatic cancer cells while inhibiting cell apoptosis.18 Moreover, miR-424-5p was upregulated in gastric cancer, and its overexpression promoted the proliferation of gastric cancer cells.19 Thus, miRNA-424-5p can play completely opposite roles in different cancers. However, the role of miRNA-424-5p in LSCC remains unknown.
In this study, we evaluated the expression of miRNA 424-5p in LSCC tissues by qPCR and analyzed the association of miR-424-5p level and clinical features of LSCC. Furthermore, we used target prediction and functional annotation and explored the role of miR-424-5p in regulating LSCC by molecular and cellular experiments.
Materials and Methods
Tissue Samples and Ethical Statement
LSCC tissues and paired adjacent normal mucosa (ANM) tissues were collected from 106 LSCC patients undergoing surgery in the Department of Otolaryngology – Head and Neck Surgery, First Hospital of Shanxi Medical University, between January 2017 and August 2018. Patients did not receive chemoradiotherapy or other biological therapies before surgery. The dates of laryngeal cancer surgery and last follow-up and status at last follow-up (living, lost to follow-up or deceased) were recorded. The diagnosis of laryngeal cancer from paraffin-embedded tissues was provided by two pathologists. All patients signed informed consent, acknowledging that they understood their rights and obligations. The study was approved by the Research Ethics Committee of Shanxi Medical University.
Microarray Data Analysis
Our previous study investigated the miRNA expression profiles of six LSCC tissues and paired ANM tissues by miRNA microarray analysis.20 After screening differentially expressed miRNAs by using the LIMMA package,21 the miRNAs with |log2 FC (fold change)| > 1, adjusted p-value < 0.01, and differentially expressed in both lymphatic metastasis and LSCC tissue with no lymphatic metastasis underwent hierarchical clustering with the pheatmap package [https://cran.r-project.org/web/packages/pheatmap/].
Bioinformatics Analysis
Target gene prediction of miR-424-5p involved using the online tool ENCORI,22 then Venn analysis was performed to obtain common target genes predicted by TargetScan, PITA, miRanda and PicTar. Subsequently, common target genes underwent Gene Ontology (GO) and KEGG pathway analysis by using DAVID 6.8.23 Transcriptome sequencing data for 493 head and neck squamous cell carcinoma samples from The Cancer Genome Atlas (TCGA) database were used for survival analysis; 111 were LSCC samples. Patients were divided into low and high expression groups based on median miR-424-5p expression for survival analysis.
RNA Extraction and qPCR Analysis
Total RNA was extracted from tissue or cells by using TRIzol reagent (Invitrogen). Reverse transcription of miRNA and mRNA involved using the All-in-One miRNA First-Strand cDNA Synthesis Kit (GeneCopoeia, Rockville, MD, USA) and HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). qPCR involved a StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA). The procedures for qPCR were 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C for 30. The 2−ΔΔct method was used to calculate the relative expression of target genes. 18S rRNA was the internal control. The primer sequences were as follows:
miR-424-5p F: 5ʹ-CAGCAGCAATTCATGTTTTGA-3ʹ;
18S RNA-F: 5ʹ-CCTGGATACCGCAGCTAGGA-3ʹ;
18S RNA-R: 5ʹ-GCGGCGCAATACGAATGCCCC-3ʹ;
CADM1-F: 5ʹ-CCCGTACAGGAGAAGAAAGAAG-3ʹ;
CADM1-R: 5ʹ-GATACTTGCTGGGTGACTACTG-3ʹ.
Cell Culture and Transfection
The LSCC cell line FD-LSC-1 was a gift from Professor Liang Zhou (Department of Otolaryngology – Head and Neck Surgery, Eye, Ear, Nose and Throat Hospital, Fudan University)24 and cells were cultured in BEGM Medium (Lonza, Walkersville, MD, USA) supplemented with 10% fetal bovine serum (FBS; BI, Cromwell, CT) and 1% L-glutamine (Gibco, Grand Island, NY). The LSCC cell line TU177 (Bioleaf Biotech Co., Shanghai) was cultured in MEM supplemented with 10% FBS. miR-424-5p mimics and negative control mimics (NC mimics) were from GenePharma (Shanghai). Cells were transfected with miR-424-5p or NC mimics at a final concentration of 30 nM by using Lipofectamine 3000 reagent according to the manufacturer’s instructions.
Cell Proliferation Assay
Cell proliferation was determined by using the Cell Counting Kit-8 (CCK8) (Transgen, Beijing) after transfection for 24, 48, 72, and 96 hrs. Moreover, EdU staining was used to assess proliferation with the Cell-Light EdU imaging kit (RiboBio Co., Guangzhou, China) following the manufacturer’s instructions.
Colony-Formation Assay
Cells were seeded in 96-well plates at 200/well and cultured for 7 days, then rinsed with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde fixation for 15 mins. Cells were stained with 0.1% crystal violet, washed with flow water, then air-dried and photographed.
Wound Healing Migration Assay
FD-LSC-1 or TU-177 cells were plated in culture-insert 2 wells, and wound healing migration assay followed the manufacturer’s instructions (ibidi, Fitchburg, WI). BEGM medium (for FD-LSC-1 cells) or MEM medium (for TU-177 cells) supplemented with 1% FBS was added, and images of scratch closure were captured at 0 and 24 hrs.
Transwell Migration Assay
After transfection, 1×106 cells were plated in 100 μL medium without serum and incubated in Transwell plates at 37°C and 5% CO2 for 24 hrs. Then, the upper side of the filter membrane was wiped with a cotton swab to remove cell debris. Cells on the lower side of the insert filter were stained with crystal violet for 10 mins and washed with PBS once.
Transwell Invasion Assay
Matrigel was thawed at 4°C, then 40 μL Matrigel solution (Matrigel:medium=1:3) was added to precooled Transwell inserts and solidified in a 37°C incubator for 2 hrs. Cells were resuspended with serum-free medium at 1 x 105 cells/well and seeded. The upper side of the filter membrane was wiped with a cotton swab to remove cell debris, then cells on the lower side of the insert filter were stained with crystal violet for 10 mins and washed with PBS once.
Cell Adhesion Assay
We precoated 96-well cell culture plates with Matrigel (2 μg/well) overnight at room temperature. FD-LSC-1 and TU-177 cells were transfected with miR-424-5p mimics and NC mimics for 24 hrs, then harvested and plated on precoated 96-well plates at 1× 104 cells/well with serum-free medium and incubated for the indicated time at 37°C. The medium containing unattached cells was removed and cells were washed once with PBS, then fixed with 4% formaldehyde and stained with 0.1% crystal violet. The dye was solubilized with 33% acetic acid, and the absorption at 595 nm was determined by using a microplate reader Multiscan MK3 (ThermoFisher Scientific).
Cell Cycle Analysis
About 1×106 cells were harvested at 48 hrs after transfection and fixed overnight in 70% ice-cold ethanol and 4℃. Cells were stained with 50 μg/mL propidium iodide, 100 U/mL RNase A, and 0.2% Triton X-100 for 30 mins, then quantified by NovoCyte flow cytometry (ACEA Biosciences, Hangzhou, China), and data were analyzed by using NovoExpress software.
Plasmid Construction and Luciferase Reporter Assay
The wild-type CADM1 3ʹ UTR luciferase reporter was generated by inserting the PCR-amplified 3ʹUTR sequence containing the predicted miR-424-5p binding site into the psiCHECK-2 vector (Promega, Madison, WI). The mutant CADM1 3ʹUTR luciferase reporter was produced by overlapping extension PCR. Primers used for luciferase reporter plasmids construction were as follows: CADM1 WT-F: 5ʹ-TCGCTCGAGTATTTCCTCCCTCTACCCCAATT-3ʹ, CADM1 WT-R: 5ʹ-CGGGAATTCCCTCCCTACTTCCCCTCCTGT-3ʹ, CADM1 Mut-F: 5ʹ-AACAATGAAAACTCTTACGTCGAGCCCATGTTTCAAAT-3ʹ, CADM1 Mut-R: 5ʹ-ATTTGAAACATGGGCTCGACGTAAGAGTTTTCATTGTT-3ʹ. Luciferase reporter assays followed the manufacturer’s instructions (Promega). Briefly, cells were seeded in 24-well plates and co-transfected with NC or miR-424-5p mimics and WT or mutant CADM1 3ʹ UTR reporter plasmid (0.1 μg/well). At 48 hrs after transfection, cells were washed with PBS, then lysed with 250 μL passive lysis buffer. Cell lysates were centrifuged at 12000 rpm for 5 mins. An amount of 50 μL supernatant was transferred to 96-well plates, injected with luciferase substrates, and luciferase activity was measured on a SpectraMax i3x Multi-Mode Microplate Reader (Molecular Devices, San Jose, CA).
Western Blot Analysis
Cells were washed twice with PBS, and protein was harvested by using RIPA reagent containing protease inhibitor cocktail (ThermoFisher Scientific). Protein concentration was measured by using the Coomassie (Bradford) Protein Assay Kit (ThermoFisher Scientific) according to the manufacturer’s instructions. Equal quantities of protein were separated by 12% SDS-PAGE and transferred to PVDF membranes (Millipore, Bedford, MA), which were blocked with 5% skim milk in TBST for 2 hrs at room temperature, then incubated with primary antibodies overnight at 4°C, followed by a TBST wash and secondary antibody incubation for 2 hrs at room temperature. Finally, membranes were incubated with Western LumaxLight Superior HRP substrate (ZETA LIFE Inc.) and protein bands were captured on a MiniChemi I instrument (SageCreation, Beijing). The primary antibodies used were anti-CADM1 (Proteintech, Cat#14335-1-AP) and anti-GAPDH (TransGen Biotec, Cat#HC301-02).
Statistical Analysis
Statistical analysis involved using SPSS v19.0 (SPSS Inc., Chicago, IL). Independent Student t test was used to compare the differences between the two groups. The difference in relative level of miR-424-5p by tumor-node-metastasis (TNM) staging and differentiation of LSCC involved the Mann–Whitney U-test. NC mimics group in all experiments was performed three times as the miR-424-5p mimics group, and the fold change in the miR-424-5p mimics group was normalized to the NC mimics group. P<0.05 was considered statistically significant.
Results
Upregulation of miR-424-5p in LSCC Is Associated with Aggressive Clinical Features of LSCC
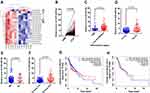
Recently, we investigated the miRNA expression profile of 6 LSCC and paired ANM tissues by microarray analysis. Several miRNAs were upregulated in LSCC versus ANM tissue. miR-424-5p was upregulated in LSCC for each pair of tissues (Figure 1A). To validate this result, we enrolled 106 patients with LSCC to measure the expression of miR-424-5p in LSCC and ANM tissues by qPCR; clinical features of these patients are shown in Table 1. qPCR results confirmed that the expression of miR-424-5p was significantly upregulated in LSCC tissue as compared with ANM tissue (Figure 1B).
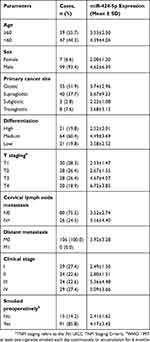 |
Table 1 Clinical Features and Relative Expression of miR-424-5p of 106 Laryngeal Squamous Cell Carcinoma (LSCC) Samples |
Next, we analyzed the association of miR-424-5p level with clinical features of LSCC patients. High miR-424-5p expression was significantly associated with poor differentiation of LSCC (Figure 1C, P=0.028 between high vs low and medium groups). Furthermore, miR-424-5p level was associated with advanced T stage and cervical lymph node metastasis (Figure 1D and E). The expression of miR-424-5p in LSCC was significantly higher with high than low clinical stage (Figure 1F, P<0.001). Moreover, survival analysis with data from the TCGA database showed high expression of miR-424-5p associated with poor prognosis with head and neck squamous cell carcinoma (HNSCC) (Figure 1G) and LSCC (Figure 1H), although not significantly. Thus, miR-424-5p was upregulated in LSCC tissue and associated with clinical features of LSCC, which indicates its critical role in the occurrence and progression of LSCC.
Functional Annotation of miR-424-5p Target Genes
To understand the potential functional role of miR-424-5p, we predicted miR-424-5p target genes by using TargetScan, miRanda, PITA, and Pictar. Venn analysis showed 492 genes associated with miR-424-5p (Figure 2A, Supplementary Table 1). On GO and KEGG pathway enrichment analysis, miR-424-5p target genes were involved in biological processes such as cell division, cell cycle, and cell migration (Figure 2B). KEGG pathway analysis showed that miR-424-5p target genes were mainly enriched in pathways in cancer, PI3K-Akt signaling pathway, signaling pathways regulating pluripotency of stem cells, cell cycle, Wnt signaling pathway, p53 signaling pathway, and several cancer-related pathways (Figure 2C). These data indicate that miR-424-5p may play important roles in the malignant behavior of cancer by regulating the expression of target genes involved in these pathways.
Overexpression of miR-424-5p Promotes the Proliferation of LSCC Cells
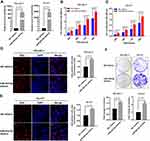
To investigate the biological roles of miR-424-5p in LSCC progression, LSCC cell lines FD-LSC-1 and TU-177 cells were transfected with miR-424-5p mimics or negative control mimics (NC mimics). The expression of miR-424-5p in FD-LSC-1 and TU-177 cells was highly increased as compared with controls after miR-424-5p mimics transfection (Figure 3A). Overexpression of miR-424-5p promoted the proliferation of FD-LSC-1 and TU-177 cells (Figure 3B and C). Moreover, the EdU-positive cell amount was significantly higher with miR-424-5p than NC mimics, so high miR-424-5p level promoted LSCC cell proliferation (Figure 3D and E). In addition, overexpression of miR-424-5p increased the colony-formation ability of LSCC cells (Figure 3F). Collectively, these results suggest that overexpression of miR-424-5p promoted LSCC cell proliferation.
Overexpression of miR-424-5p Promotes Migration and Invasion of LSCC Cells
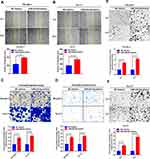
Invasion and cervical lymph node metastasis are important characteristics of LSCC.25 To evaluate the effects of miR-424-5p on the invasion and migration of LSCC cells, we used scratch-wound healing and transwell assays with FD-LSC-1 and TU-177 cells transfected with miR-424-5p or NC mimics. As compared with NC mimics, with miR-424-5p mimics, the distance of migration of cells was increased significantly (Figure 4A and B). Transwell migration assay confirmed that overexpression of miR-424-5p promoted LSCC cell migration (Figure 4C).
Cells transfected with miR-424-5p mimics showed enhanced invasive ability as compared with NC mimics (Figure 4D). Highly metastatic cancer cells show enhanced adhesion ability that facilitates the migration of the cells to a new site to establish new tumors.26 Therefore, cell adhesion assay is used to evaluate the metastatic ability of LSCC cells. We found that overexpression of miR-424-5p significantly increased adhesive ability as compared with NC group (Figure 4E and F). Therefore, overexpression of miR-424-5p enhanced the migration and invasion of LSCC cells.
Overexpression of miR-424-5p Affects the Cell Cycle Progression of LSCC Cells
Induction of cell cycle acceleration is an important mechanism for promoting cell proliferation.27 Here, we analyzed the cell cycle population in FD-LSC-1 and TU-177 cells with miR-424-5p overexpression by flow cytometry. The number of cells in the S and G2 phases was significantly increased after transfection with miR-424-5p mimics than the NC mimics, and the number of cells in the G1 phase was decreased (Figure 5A and B).
CADM1 as a Direct Target of miR-424-5p in LSCC Cells
To further elucidate the potential mechanism of how miR-424-5p promotes the malignant behavior of LSCC cells, we validated the regulatory effect of miR-424-5p on the predicted target gene CADM1. Overexpression of miR-424-5p significantly downregulated CADM1 mRNA level (Figure 6A) and CADM1 protein level was reduced in LSCC cells transfected with miR-424-5p or NC mimic (Figure 6B). We further investigated the regulatory effect of miR-424-5p on CADM1 by luciferase reporter assay. The luciferase activity of wild-type CADM1 3ʹUTR luciferase reporter was significantly reduced in miR-424-5p–overexpressing cells, but the mutant CADM1 3ʹUTR luciferase did not respond to miR-424-5p overexpression (Figure 6C). Collectively, these results suggest that miR-424-5p targets CADM1 to suppress its expression in LSCC cells.
Discussion
Because of the difficulty in early diagnosis and poor prognosis, LSCC has become one of the rapidly growing causes of cancer death worldwide. The miRNAs involved in carcinogenesis due to their oncogenic or tumor-suppressive functions and recent studies revealed that miRNAs play crucial roles in LSCC.11,28,29 We and other groups have investigated the gene expression profiles of LSCC tissues and found a number of miRNAs dysregulated in LSCC.10,20 Thus, further exploring the function and clinical significance of differentially expressed miRNAs is a key step for developing new biomarkers and targets for LSCC. In the current study, we found miR-424-5p was upregulated in LSCC versus ANM tissues. High miR-424-5p level was significantly associated with poor differentiation, advanced tumor stage and cervical lymph node metastasis. miR-424-5p target genes are mainly enriched in biological processes of the cell cycle, cell division, and negative regulation of cell migration and were involved in multiple cancer-related pathways. Overexpression of miR-424-5p promoted proliferation, migration, invasion, and adhesion of LSCC cells. Furthermore, CADM1 was a direct target of miR-424-5p in LSCC cells.
Emerging evidence has revealed that miR-424-5p has oncogene and tumor suppressor roles in different cancers. Expression of miR-424-5p was downregulated in epithelial ovarian cancer and hepatocellular carcinoma tissues, and its overexpression inhibited cell proliferation in these cancers.16,30 However, Wei et al reported that miR-424-5p was upregulated in gastric cancer tissues, and restoration of miR-424-5p could promote the proliferation of gastric cancer cells.19 The different miR-424-5p expression patterns in various cancers may be result of differential DNA methylation level, transcription and post-transcriptional regulation. Hypermethylation of the miR-424 promoter region has been reported in glioma and cervical cancer tissues, and treatment of glioma and cervical cancer cell lines with the demethylation reagent 5-aza-2ʹ-deoxycytidine increased miR-424-5p expression,31,32 so epigenetic repression by promoter region hypermethylation may be an important mechanism of downregulation of miR-424-5p in some cancers. By contrast, upregulation of miR-424-5p in cancers involves more complex transcriptional and post-transcriptional mechanisms, such as dysregulated upstream transcription factors and long non-coding RNA.33–35
In the present study, miR-424-5p was significantly upregulated in LSCC. Importantly, high expression of miR-424-5p was significantly associated with poor differentiation, advanced T stage and clinical stage. Clinical features associated with miRNAs play important regulatory roles in cancer progression, including miR-145-5p, miR-195 and miR-106b-5p.11,36,37 Hence, we speculated that miR-424-5p is a critical regulator of LSCC.
Cancer cells have enhanced adhesion capacity and viability38–40 and abnormal activation of PI3K-Akt signaling pathway.41,42 Moreover, most recent studies concluded that cancer stem cells are the origin of proliferation, migration and chemoresistance of cancer cells.43–45 Our previous study isolated and characterized cancer stem cells from LSCC cells.46,47 In the current study, GO and KEGG analysis revealed that potential target genes of miR-424-5p were enriched in biological processes and signaling pathways of cell adhesion, cell division, cell cycle, and cell migration, PI3K-Akt signaling pathway, signaling pathways regulating pluripotency of stem cells, as well as several cancer-related pathways, so miR-424-5p may affect malignant phenotypes of LSCC through these signaling pathways.
However, to date, no study has reported the function of miR-424-5p in LSCC cells. FD-LSC-1 and TU-177 are among the few well-characterized human LSCC cell lines,24,48 hence we used these two cell lines as model to investigate the role of miR-424-5p in LSCC cells. Our results showed that overexpression of miR-424-5p significantly promoted the proliferation of the LSCC cells. Furthermore, restoration of miR-424-5p expression enhanced the migration, invasion, and adhesion of LSCC cells. These results were similar to findings of miR-424-5p function in pancreatic cancer and non-small-cell lung cancer cells.18,49 Moreover, we found that overexpression of miR-424-5p promoted the cell cycle transition from the G1 phase to S and G2/M phases. This finding agrees with the previous study in esophageal squamous cell carcinoma cells33 and may explain in part the proliferation-promoting role of miR-424-5p on LSCC cells. Collectively, these data suggest that miR-424-5p plays an oncogenic role in LSCC to promote proliferation, migration, invasion, adhesion, and cell cycle progression.
Furthermore, our data revealed that miR-424-5p suppresses the expression of CADM1. CADM1 is a well-known tumor suppressor, downregulation of CADM1 was found in hepatocellular carcinoma,50,51 breast cancer,52 and melanoma.53 Restoration of CADM1 level inhibited proliferation, invasion, and tumorigenesis; negatively regulated G1/S transition; and promoted apoptosis of cancer cells.50,51,54–56 Additionally, downregulation of CADM1 level-enhanced chemoresistance in tongue cancer cells.57 Therefore, CADM1 may be a critical downstream target of miR-424-5p in LSCC and miR-424-5p may promote LSCC progression by downregulating CADM1 level.
Notably, we investigated the function of miR-424-5p based on in vitro experiments. Considering that in vitro studies lack the information of the mutual interaction between cancer cells and the microenvironment, in the future, in vivo experiments using xenograft tumors are needed to clarify the functional role of miR-424-5p in LSCC.
Conclusion
Our data support that miR-424-5p may promote the proliferation, migration, invasion, and adhesion of LSCC cells, and upregulation of miR-424-5p is closely associated with malignant clinical features of LSCC in humans. Furthermore, the tumor suppressor CADM1 is a direct target of miR-424-5p. miR-424-5p may be a promising therapeutic target and a potential diagnostic biomarker for LSCC.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 81602394, 81802793, 81802948, 81872210), Shanxi Province Scientific and Technological Achievements Transformation Guidance Foundation (Nos. 201604D131002 and 201604D132040), Key Technological Innovation Platform Foundation for Head and Neck Cancer Research of Shanxi Province (No. 201605D151003), The Research Project of Shanxi Province Health and Family Planning Commission (No. 201601038), the Excellent Talent Science and Technology Innovation Project of Shanxi Province (Nos. 201705D211018, 201805D211007), Natural Science Foundation of Basic Research Program of Shanxi Province (Nos. 201801D221421, 201801D221428), Outstanding Youth Development Foundation of The First Hospital Affiliated with Shanxi Medical University (No. YR1601), and Startup Foundation for Doctors of Shanxi Medical University (No. BS03201624).
Disclosure
The authors report no conflict of interest in this work.
References
1. Steuer CE, El-Deiry M, Parks JR, Higgins KA, Saba NF. An update on larynx cancer. CA Cancer J Clin. 2017;67(1):31–50. doi:10.3322/caac.21386
2. Gao W, An C, Xue X, et al Mass spectrometric analysis identifies AIMP1 and LTA4H as FSCN1 binding proteins in laryngeal squamous cell carcinoma. Proteomics. 2019;19:e1900059. doi:10.1002/pmic.v19.21-22
3. Chen P, Yu W, Huang J, et al Matched-pair analysis of survival in patients with poorly differentiated versus well-differentiated glottic squamous cell carcinoma. Oncotarget. 2017;8(9):14770–14776. doi:10.18632/oncotarget.14772
4. Zhao X, Zhang W, Ji W. YB-1 promotes laryngeal squamous cell carcinoma progression by inducing miR-155 expression via c-Myb. Future Oncol. 2018;14(16):1579–1589. doi:10.2217/fon-2018-0058
5. Byzia E, Soloch N, Bodnar M, et al Recurrent transcriptional loss of the PCDH17 tumor suppressor in laryngeal squamous cell carcinoma is partially mediated by aberrant promoter DNA methylation. Mol Carcinog. 2018;57(7):878–885. doi:10.1002/mc.v57.7
6. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
7. Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol Biol. 2017;1509:1–10.
8. Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: new trends in the development of miRNA therapeutic strategies in oncology (Review). Int J Oncol. 2016;49(1):5–32. doi:10.3892/ijo.2016.3503
9. Janiszewska J, Szaumkessel M, Kostrzewska-Poczekaj M, et al Global miRNA expression profiling identifies miR-1290 as novel potential oncomir in laryngeal carcinoma. PLoS ONE. 2015;10(12):e0144924. doi:10.1371/journal.pone.0144924
10. Cao P, Zhou L, Zhang J, et al Comprehensive expression profiling of microRNAs in laryngeal squamous cell carcinoma. Head Neck. 2013;35(5):720–728. doi:10.1002/hed.v35.5
11. Gao W, Zhang C, Li W, et al Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol Ther. 2019;27(2):365–379. doi:10.1016/j.ymthe.2018.09.018
12. Gao W, Wu Y, He X, et al MicroRNA-204-5p inhibits invasion and metastasis of laryngeal squamous cell carcinoma by suppressing forkhead box C1. J Cancer. 2017;8(12):2356–2368. doi:10.7150/jca.19470
13. Lu Y, Gao W, Zhang C, et al Hsa-miR-301a-3p acts as an oncogene in laryngeal squamous cell carcinoma via target regulation of Smad4. J Cancer. 2015;6(12):1260–1275. doi:10.7150/jca.12659
14. Lian R, Lu B, Jiao L, et al MiR-132 plays an oncogenic role in laryngeal squamous cell carcinoma by targeting FOXO1 and activating the PI3K/AKT pathway. Eur J Pharmacol. 2016;792:1–6. doi:10.1016/j.ejphar.2016.10.015
15. Xu J, Li Y, Wang F, et al Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–987. doi:10.1038/onc.2012.121
16. Liu J, Gu Z, Tang Y, Hao J, Zhang C, Yang X. Tumour-suppressive microRNA-424-5p directly targets CCNE1 as potential prognostic markers in epithelial ovarian cancer. Cell Cycle. 2018;17(3):309–318. doi:10.1080/15384101.2017.1407894
17. Zhang Y, Li T, Guo P, et al MiR-424-5p reversed epithelial-mesenchymal transition of anchorage-independent HCC cells by directly targeting ICAT and suppressed HCC progression. Sci Rep. 2014;4:6248.
18. Wu K, Hu G, He X, et al MicroRNA-424-5p suppresses the expression of SOCS6 in pancreatic cancer. Pathol Oncol Res. 2013;19(4):739–748. doi:10.1007/s12253-013-9637-x
19. Wei S, Li Q, Li Z, Wang L, Zhang L, Xu Z. miR-424-5p promotes proliferation of gastric cancer by targeting Smad3 through TGF-β signaling pathway. Oncotarget. 2016;7(46):75185–75196. doi:10.18632/oncotarget.12092
20. Zhang C, Gao W, Wen S, et al Potential key molecular correlations in laryngeal squamous cell carcinoma revealed by integrated analysis of mRNA, miRNA and lncRNA microarray profiles. Neoplasma. 2016;63(6):888–900. doi:10.4149/neo_2016_608
21. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi:10.2202/1544-6115.1027
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248
23. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
24. Wu CP, Zhou L, Gong HL, et al Establishment and characterization of a novel HPV-negative laryngeal squamous cell carcinoma cell line, FD-LSC-1, with missense and nonsense mutations of TP53 in the DNA-binding domain. Cancer Lett. 2014;342(1):92–103. doi:10.1016/j.canlet.2013.08.041
25. Lucioni M, D’Ascanio L, De Nardi E, Lionello M, Bertolin A, Rizzotto G. Management of paratracheal lymph nodes in laryngeal cancer with subglottic involvement. Head Neck. 2018;40(1):24–33. doi:10.1002/hed.v40.1
26. Chen Y, Lu B, Yang Q, Fearns C, Yates JR, Lee JD. Combined integrin phosphoproteomic analyses and small interfering RNA-based functional screening identify key regulators for cancer cell adhesion and migration. Cancer Res. 2009;69(8):3713–3720. doi:10.1158/0008-5472.CAN-08-2515
27. Ren TN, Wang JS, He YM, Xu CL, Wang SZ, Xi T. Effects of SMYD3 over-expression on cell cycle acceleration and cell proliferation in MDA-MB-231 human breast cancer cells. Med Oncol. 2011;28(Suppl 1):S91–98. doi:10.1007/s12032-010-9718-6
28. Saito K, Inagaki K, Kamimoto T, et al MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS ONE. 2013;8(8):e71480. doi:10.1371/journal.pone.0071480
29. Gao W, Zhang Y, Niu M. et al Identification of miR-145-5p-centered competing endogenous RNA network in laryngeal squamous cell carcinoma. Proteomics;2019. e1900020. doi:10.1002/pmic.201900020
30. Du H, Xu Q, Xiao S, et al MicroRNA-424-5p acts as a potential biomarker and inhibits proliferation and invasion in hepatocellular carcinoma by targeting TRIM29. Life Sci. 2019;224:1–11. doi:10.1016/j.lfs.2019.03.028
31. Jin C, Li M, Ouyang Y, Tan Z, Jiang Y. MiR-424 functions as a tumor suppressor in glioma cells and is down-regulated by DNA methylation. J Neurooncol. 2017;133(2):247–255. doi:10.1007/s11060-017-2438-4
32. Varghese VK, Shukla V, Kabekkodu SP, Pandey D, Satyamoorthy K. DNA methylation regulated microRNAs in human cervical cancer. Mol Carcinog. 2018;57(3):370–382. doi:10.1002/mc.v57.3
33. Wen J, Hu Y, Liu Q, et al miR-424 coordinates multilayered regulation of cell cycle progression to promote esophageal squamous cell carcinoma cell proliferation. EBioMedicine. 2018;37:110–124. doi:10.1016/j.ebiom.2018.10.043
34. Drasin DJ, Guarnieri AL, Neelakantan D, et al TWIST1-induced miR-424 reversibly drives mesenchymal programming while inhibiting tumor initiation. Cancer Res. 2015;75(9):1908–1921. doi:10.1158/0008-5472.CAN-14-2394
35. Zhou K, Li S, Du G, et al LncRNA XIST depletion prevents cancer progression in invasive pituitary neuroendocrine tumor by inhibiting bFGF via upregulation of microRNA-424-5p. Onco Targets Ther. 2019;12:7095–7109. doi:10.2147/OTT.S208329
36. Li B, Wang S, Wang S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol Genet Genomics. 2018;293(5):1245–1253. doi:10.1007/s00438-018-1457-y
37. Shi DM, Bian XY, Qin CD, Wu WZ. miR-106b-5p promotes stem cell-like properties of hepatocellular carcinoma cells by targeting PTEN via PI3K/Akt pathway. Onco Targets Ther. 2018;11:571–585. doi:10.2147/OTT.S152611
38. van de Merbel AF, van der Horst G, Buijs JT, van der Pluijm G. Protocols for migration and invasion studies in prostate cancer. Methods Mol Biol. 2018;1786:67–79.
39. Boege Y, Malehmir M, Healy ME, et al A dual role of Caspase-8 in triggering and sensing proliferation-associated DNA damage, a key determinant of liver cancer development. Cancer Cell. 2017;32(3):342–359.e10. doi:10.1016/j.ccell.2017.08.010
40. Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29(6):783–803. doi:10.1016/j.ccell.2016.05.005
41. Guo H, German P, Bai S, et al The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42(7):343–353. doi:10.1016/j.jgg.2015.03.003
42. Chen H, Zhou L, Wu X, et al The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front Biosci (Landmark Ed). 2016;21:1084–1091. doi:10.2741/4443
43. Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol. 2015;31:28–35. doi:10.1016/j.semcancer.2014.07.001
44. Li Y, Atkinson K, Zhang T. Combination of chemotherapy and cancer stem cell targeting agents: preclinical and clinical studies. Cancer Lett. 2017;396:103–109. doi:10.1016/j.canlet.2017.03.008
45. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi:10.1038/nm.4409
46. Wu Y, Zhang Y, Niu M, et al Whole-transcriptome analysis of CD133+CD144+ cancer stem cells derived from human laryngeal squamous cell carcinoma cells. Cell Physiol Biochem. 2018;47(4):1696–1710. doi:10.1159/000490992
47. Wang J, Wu Y, Gao W, et al Identification and characterization of CD133+CD44+ cancer stem cells from human laryngeal squamous cell carcinoma cell lines. J Cancer. 2017;8(3):497–506. doi:10.7150/jca.17444
48. Liu TJ, Zhang WW, Taylor DL, Roth JA, Goepfert H, Clayman GL. Growth suppression of human head and neck cancer cells by the introduction of a wild-type p53 gene via a recombinant adenovirus. Cancer Res. 1994;54(14):3662–3667.
49. Zhang M, Gao C, Yang Y, et al MiR-424 promotes non-small cell lung cancer progression and metastasis through regulating the tumor suppressor gene TNFAIP1. Cell Physiol Biochem. 2017;42(1):211–221. doi:10.1159/000477314
50. Zhang W, Xie HY, Ding SM, et al CADM1 regulates the G1/S transition and represses tumorigenicity through the Rb-E2F pathway in hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2016;15(3):289–296. doi:10.1016/S1499-3872(16)60099-1
51. Sun Z, Meng C, Wang S, et al MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC Cancer. 2014;14:616. doi:10.1186/1471-2407-14-616
52. Wikman H, Westphal L, Schmid F, et al Loss of CADM1 expression is associated with poor prognosis and brain metastasis in breast cancer patients. Oncotarget. 2014;5(10):3076–3087. doi:10.18632/oncotarget.v5i10
53. Mou K, Zhang X, Mu X, et al LNMAT1 promotes invasion-metastasis cascade in malignant melanoma by epigenetically suppressing CADM1 expression. Front Oncol. 2019;9:569. doi:10.3389/fonc.2019.00569
54. Zhang G, Zhong L, Luo H, Wang S. MicroRNA-155-3p promotes breast cancer progression through down-regulating CADM1. Onco Targets Ther. 2019;12:7993–8002. doi:10.2147/OTT.S206180
55. Wang HL, Zhou R, Liu J, et al MicroRNA-196b inhibits late apoptosis of pancreatic cancer cells by targeting CADM1. Sci Rep. 2017;7(1):11467. doi:10.1038/s41598-017-11248-3
56. Vallath S, Sage EK, Kolluri KK, et al CADM1 inhibits squamous cell carcinoma progression by reducing STAT3 activity. Sci Rep. 2016;6:24006. doi:10.1038/srep24006
57. Zheng G, Li N, Jia X, et al MYCN-mediated miR-21 overexpression enhances chemo-resistance via targeting CADM1 in tongue cancer. J Mol Med (Berl). 2016;94(10):1129–1141. doi:10.1007/s00109-016-1417-0
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.