Back to Journals » OncoTargets and Therapy » Volume 9
MiR-381 functions as a tumor suppressor in colorectal cancer by targeting Twist1
Authors He X, Wei Y, Wang Y, Liu L, Wang W, Li N
Received 27 October 2015
Accepted for publication 16 December 2015
Published 7 March 2016 Volume 2016:9 Pages 1231—1239
DOI https://doi.org/10.2147/OTT.S99228
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
Xinxin He, Yangnian Wei, Yong Wang, Ling Liu, Wen Wang, Nianfeng Li
Department of Hepatobiliary and Pancreatic Surgery, Xiangya Hospital, Central South University, Hunan, People’s Republic of China
Abstract: MiR-381 has been reported to be dysregulated in several human cancers. However, the function and mechanism of miR-381 in colorectal cancer (CRC) remains unclear. In the present study, the miR-381 expression was assessed in a cohort of 113 CRC specimens using real-time quantitative polymerase chain reaction (RTq-PCR), which demonstrated that miR-381 was significantly downregulated in CRC and correlated with distant metastasis and tumor, node, and metastasis (TNM) stage. Functional study revealed that restoration of miR-381 significantly inhibited the invasion, migration, and epithelial–mesenchymal transition (EMT) of CRC cells. Luciferase reporter assay validated that Twist1, an important EMT inducer, was a direct target of miR-381, and rescued Twist1 attenuated the function of miR-381 in CRC cells. Correlation analysis also revealed an inverse correlation between miR-381 and Twist1 expression levels in CRC specimens. Taken together, our results highlight the significance of miR-381/Twist1 interaction in the development and progression of CRC, and suggest that restoration of miR-381 may be a potential therapeutic strategy for the patients with CRC.
Keywords: colorectal cancer, miR-381, twist family bHLH transcription factor 1, Twist1, epithelial–mesenchymal transition,
EMT
Introduction
Colorectal cancer (CRC) is one of the most malignant and leading causes of cancers worldwide.1,2 Although there has been substantial progress in the treatment for patients with CRC, approximately half of them die within 5 years, mainly due to metastasis;3,4 thus, further investigation into the underlying molecular mechanisms is of great clinical significance. Recently, increasing reports have revealed the close relationship between dysregulated microRNAs (miRNAs) and the development of CRC, which highlights the role of miRNAs as potential biomarkers and therapeutic targets in patients with CRC.5–7
miRNAs are a class of short and noncoding RNAs that negatively regulate the expression of protein-coding genes by directly binding their 3′ untranslated region (3′-UTR), leading to the posttranscriptional translation inhibition or mRNA degradation.8,9 In CRC, many miRNAs have been found to be involved in the regulation of multiple cellular functions, such as cell proliferation, invasion, and migration.10 For example, miR-143 and miR-145 function as tumor suppressors by inhibiting cell proliferation, invasion, and metastasis,11,12 while miR-21 acts as an important promoter of oncogenesis.13,14 CRC pathogenesis is particularly accompanied by the epithelial–mesenchymal transition (EMT) that is considered the primary step in distant metastasis of many types of cancer, the major concern in clinical cancer therapy.15,16 Recent findings showed that miRNA is important in regulating the EMT process in CRC.17–19 Therefore, identification of EMT-associated miRNAs as biomarkers or therapeutic targets is of great importance.
Decreased miR-381 has been reported in breast,20 pituitary,21 and prostate cancer,22 suggesting its tumor suppressive function. For example, Ming et al found miR-381 suppressed the migration and invasion of breast cancer cells by targeting Cx43, an enhancer of the metastasis of breast cancer cells.20 Recently, Liang et al also confirmed that miR-381 was downregulated in colon cancer.23 However, the clinical significance and role of miR-381 in CRC remains unclear. In the present study, we examined the expression levels of miR-381 in an expanded cohort of CRC tissues and several cell lines and found that miR-381 was significantly downregulated in CRC tissues and significantly correlated with distant metastasis and tumor, node, and metastasis (TNM) stage. Further study revealed that overexpression of miR-381 could inhibit the invasiveness and EMT of CRC cells, at least partially, by targeting an important EMT inducer Twist1.
Materials and methods
Tissue samples and cell lines
A cohort of 113 CRC tumor tissues and 51 paired normal mucosal tissues were obtained from Xiangya Hospital between April 2012 and October 2014. These samples were from 113 patients with CRC who had not undergone radiation therapy or chemotherapy before the surgery and had their samples stored at −80°C. The clinicopathologic information of the patients are summarized in Table 1. Written informed consent was obtained from all the study participants and the use of these samples was approved by the ethical review committees of the Xiangya Hospital Ethic Committee of Central South University. Five CRC cell lines (HCT116, SW620, LOVO, HT29, and SW480) and normal colonic cell line NCM460 were purchased from American Type Culture Collection and cultured in suggested medium supplemented with 10% fetal bovine serum (FBS) and in 5% CO2 humid atmosphere at 37°C.
RNA isolation and RTq-PCR
Total RNA was extracted using Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instruction, and reverse transcribed using the Primer Script RT reagent Kit (TaKaRa Bio Inc., Otsu, Japan). The relative expression of Twist1 was detected by SYBR Green real-time quantitative polymerase chain reaction (RTq-PCR) assay (Bio-Rad Laboratories Inc., Hercules, CA, USA), and β-actin was used as internal control. Primers used for Twist1 and β-actin are as follows: Twist1, forward: 5′-AGAAGTCTGCGGGCTGTGGCG-3′, reverse: 5′-GAGGGCAGCGTGGGGATGATC-3′; β-actin, 5′-AGTGTGACGTGGACATCCGCAAAG-3′ (forward), 5′-ATCCACATCTGCTGGAAGGTGGAC-3′ (reverse). The relative expression of miR-381 was determined using mirVana RTq-PCR miRNA Detection Kit (Ambion, Austin, TX, USA), and small nuclear U6 RNA was used as internal control. The specific primers for miRNA-381 and U6 were purchased from GeneCopoeia (Rockville, MD, USA). All experiments were performed in at least triplicate and the relative expression levels were calculated using the 2−ΔΔCt method.
Knockdown of Twist1 by siRNA
To knockdown Twist1 expression, the siRNAs (si-Twist1, forward: 5′-GGUACAUCGACUUCCUCUAUU-3′; reverse: 5′-UAGAGGAAGUCGAUGUACCUU-3′) and negative control (si-NC, forward: 5′-UUCGACUGUACUCGACAUCTT-3′; reverse: 5′-GAUGUCGAGUACAGUCGAATT-3′) were purchased from GenePharma Company (Shanghai, People’s Republic of China). A total of 300 pmol of si-Twist1 or si-NC was transfected into HT29 and SW480 cells using Lipofectamine RNAi MAX Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Vector construction, lentivirus infection, and cell transfection
The coding sequence of Twist1 was amplified (forward primer: 5′-GAGATGATGCAGGACGTGTC-3′; reverse primer: 5′-GTGGGACGCGGACATGGACCA-3′) and cloned into pcDNA3.1 vector, and the empty pCDNA3.1 vector was used as control. Lipofectamine 2000 Reagent (Thermo Fisher Scientific) was used for cell transfection following the manufacturer’s protocol. The pre-miR-381 sequence was amplified (forward primer: 5′-CGTGAATGATAGTGAGGAAC-3′; reverse primer: 5′-GTGAACGATTTGCCACACACA-3′) and introduced into the PLKO.3G vector. Lentiviruses containing pre-miR-381 (miR-381) and negative control (miR-NC) were produced by GeneChem Company (Shanghai, People’s Republic of China). Cells were cultured to approximately 70% confluence and then added by a concentration of 2.0×105 TU/well lentiviruses containing pre-miR-381 or negative control, and RTq-PCR was performed to determine the expression levels of miR-381 after being infected for 7 days.
Cell proliferation, invasion, and migration assays
Cell proliferation was examined by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Briefly, the cell lines were plated in 96-well plates (3,000 per well), and allowed to grow for 24, 48, and 72 hours, then assessed by a colorimetric assay using MTT solution (10 mg/mL) at 570 nm. Cell invasion ability assay was performed using transwell invasion chambers coated with matrigel (BD Biosciences, San Jose, CA, USA). Cells were suspended in FBS-free medium and added to the upper chamber, while the medium containing 10% FBS was added to the lower chamber. After 24 hours of incubation, the cells remaining on the upper membrane were removed with cotton wool, whereas the cells that had invaded through the membrane were stained with methanol and 0.1% crystal violet, imaged, and counted using an inverted microscope (Olympus, Tokyo, Japan). Cell migration ability was assessed by performing wound healing assays. Cells were cultured to 100% confluence, and wounds were generated using pipette tips. The cells were then cultured for 24 or 48 hours and the wound closure was assessed by Scion Image Software (Scion Image Beta 4.03; Scion Corporation, Frederick, MD, USA).
Luciferase reporter assays
The wild-type (WT) 3′UTR of Twist1 was amplified (forward primer: 5′-TCAGAGGAACTATAAGAACACCT-3′; reverse primer: 5′-CAAGCAGGTATTTACCACCAACT-3′) and ligated into the psiCheck-2 reporter vector (Promega Corporation, Fitch-burg, WI, USA). Site-directed mutagenesis of the miR-381 seed sequence in the 3′UTR of Twist1 (Mut) was performed using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Luciferase activity was detected using the dual luciferase assay (Promega) according to the manufacturer’s protocols. Briefly, cells were cotransfected with 0.3 mg of the reporter vectors and 40 nM miR-381 mimics or scrambled mimics. Forty eight hours after transfection, the transfected cells were lysed and the relative luciferase activity was determined using a Modulus TD20/20 Luminometer (Turner Biosystems, Sunnyvale, CA, USA). All experiments were performed in triplicate. miR-381 and scramble mimics and corresponding inhibitors were purchased from RiboBio (Guangzhou, People’s Republic of China).
Western blot analysis
Total cellular extracts were prepared using 200 μL of lysis buffer. Approximately 50 μg of total protein was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to a polyvinylidene fluoride (PVDF) membrane, and incubated with the specific primary antibodies against E-cadherin, vimentin (Abgent Biotechnology, San Diego, CA, USA), N-cadherin (Cell Signaling Technology, Beverly, MA, USA), and β-actin (Santa Cruz Biotechnology Inc., Dallas, TX, USA) overnight at 4°C. Then the membranes were washed with TBST (Tris-buffered saline with Tween-20), and probed with horseradish peroxidase (HRP)-conjugated secondary antibody (1:1,500 dilution; Santa Cruz) at 37°C for 30 minutes. Signals were visualized using electrochemiluminescent (ECL) substrates (EMD Millipore, Billerica, MA, USA).
Statistical analysis
All statistical analyses were performed using the SPSS 19.0 software (SPSS Inc., Chicago, IL, USA). Experimental data were expressed as the mean ± standard deviation (SD). Statistical significance was analyzed using Student’s t-test. The correlation between miR-381 and Twist1 expression was analyzed using Spearman’s correlation analysis. P<0.05 was considered statistically significant.
Results
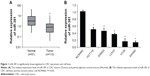
MiR-381 is downregulated in CRC specimens with distant metastasis and high TNM stages
The expression levels of miR-381 were evaluated by RTq-PCR in a cohort of 113 CRC tumor tissues and 51 paired normal mucosal tissue. As shown in Figure 1A, miR-381 expression was significantly decreased in CRC tumors compared with that in the paired normal mucosal tissue. Correlation analysis revealed that miR-381 downregulation was significantly correlated with distant metastasis (P=0.007) and high TNM stages (P=0.035), while no significant correlation was found in other clinicopathologic factors (Table 1). In addition, we also found miR-381 expression levels in CRC cell lines were significantly lower than that of the normal colonic cell line NCM460 (Figure 1B). Thus, our data suggest that decreased miR-381 expression may be associated with CRC carcinogenesis.
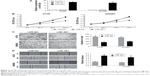
MiR-381 inhibits the proliferation, invasion, and migration of CRC cells
To investigate the functional roles of miR-381 in CRC cells, we constructed miR-381 expressing lentiviral vector and infected HT29 and SW480 cells. Overexpression of miR-381 was validated by RTq-PCR (Figure 2A). MTT assays (Figure 2B), transwell with matrigel assays (Figure 2C), and wound healing assays (Figure 2D) showed that miR-381 overexpression significantly inhibited cell proliferation, invasion, and migration ability of HT29 and SW480 cells. In contrast, when endogenous miR-381 was silenced with inhibitor mimics, cell proliferation, invasion, and migration ability was increased. These results suggest that miR-381 may be a tumor suppressor gene in CRC cells.
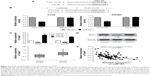
MiR-381 directly inhibits Twist1 by targeting its 3′UTR
To elucidate the molecular mechanism underlying inhibitory effects of miR-381 on proliferation, invasion, and migration of CRC cells, we predicted potential targets of miR-381. Using TargetScan and miRanda tools online, we screened several invasion- and migration-related genes by RTq-PCR, of which Twist1 expression was significantly repressed in miR-381 expressing HT29 and SW480 cells. In addition, Twist1 is an important inducer of EMT, which is involved in cell invasion and migration ability,24,25 and hence we focused on Twist1 in the study. To validate if miR-381 directly targets Twist1 3′UTR (Figure 3A), luciferase reporter assays were performed in HT29 and SW480 cells, which revealed that miR-381 mimics significantly decreased the luciferase activity of wild-type Twist1 3′UTR reporter, but had no effect on the Twist1 3′UTR reporter with mutant miR-381 binding seed sites (Figure 3B). RTq-PCR and Western blot also validated that miR-381 mimics reduced Twist1 expression, and miR-381 inhibitor mimics increased Twist1 expression in HT29 and SW480 cells (Figure 3C and D). Moreover, RTq-PCR analysis revealed that Twist1 expression was significantly increased in CRC tumors compared with paired normal mucosal tissue (Figure 3E) and inversely correlated with the expression levels of miR-381 in CRC tissues used in the study (Figure 3F). Taken together, our results demonstrated that miR-381 directly inhibits Twist1 expression in CRC cells.
Twist1 attenuates the inhibitory effects of miR-381 on the proliferation, invasion, migration, and EMT of CRC cells
We further examined whether Twist1 is a substantial target of miR-381 involved in regulating the migration and invasion of CRC cells. Knockdown of Twist1 by specific siRNAs (Figure 4A) significantly attenuated the migration and invasion of HT29 and SW480 cells (Figure 4B), similar to the effects of miR-381 overexpression (Figure 2C and D). In addition, rescued Twist1 lacking its 3′UTR in the miR-381 expressing HT29 and SW480 cells significantly attenuated inhibitory effects of miR-381 on the cell invasion and migration ability (Figure 4C). Of note, we found miR-381 significantly reversed EMT of HT29 and SW480 cells based on the observation that miR-381 increased epithelial marker (E-cadherin) and inhibited mesenchymal markers (N-cadherin and vimentin) (Figure 4D), while in Twist1 rescue experiments, we found Twist1 was a substantial EMT inducer and remarkably attenuated the inhibitory effects of miR-381 on repression of EMT in HT29 and SW480 cells (Figure 4D). Taken together, our results demonstrated that Twist1 attenuates the inhibitory effects of miR-381 on the proliferation, invasion, migration, and EMT of CRC cells.
Discussion
Increasing reports have demonstrated that miRNAs are an important regulator of tumor progression and metastasis. In CRC, many miRNAs have been identified to regulate known genes that are involved in the pathology of tumorigenesis and metastasis. In the present study, we showed that miR-381 was significantly downregulated in a cohort of 113 CRC and five CRC cell lines, which was consistent with a recently published report on miR-381 in colon cancer.23 In addition, we found decreased miR-381 expression was significantly correlated with distant metastasis and high TNM stages. These findings suggest that miR-381 may function as a tumor suppressor in CRC.
We then investigated the functional roles of miR-381 in CRC cell lines. To do so, we constructed miR-381 expressing HT29 and SW480 cells. MTT, transwell with matrigel, and wound healing assays revealed that miR-381 could inhibit cell proliferation, invasion, and migration ability of HT29 and SW480 cells. Combined with the recent observation that miR-381 inhibited the cell proliferation and invasion in colon cancer,23 we concluded that miR-381 acts as a tumor suppressor in CRC. This was also consistent with previous findings that miR-381 suppressed breast,20 pituitary,21 and prostate cancers.22 However, another report revealed that high expression of miR-381 in gliomas was involved in pathological malignant progression, suggesting miR-381 as a tumor promoter in gliomas.26 All these observations suggest that the role of miR-381 might vary in different types of cancer.
We next predicted and validated the possible targets of miR-381 in CRC cells as the impact of specific miRNAs on cancer biology depends on their downstream targets. We found Twist1 was a direct target of miR-381 by the luciferase reporter assay. To confirm the interaction of miR-381 and Twist1 in CRC, we performed RTq-PCR on Twist1 expression in CRC tissues and found a significant inverse correlation between their expression levels in clinical CRC. In addition, functional study revealed that Twist1 knockdown significantly inhibited the invasion and migration ability of HT29 and SW480 cells, and rescued Twist1 in miR-381 expressing HT29 and SW480 cells dramatically attenuated miR-381-induced inhibition on cellular migration and invasion. Taken together, our data suggest that Twist1 is a functional target of miR-381 in CRC cells.
EMT refers to the transformation of epithelial cells to mesenchymal cells, a key mechanism involved in the development of many tumors, including CRC.15,27 EMT has been associated with a more aggressive tumor phenotype including local invasion and distant metastasis.28,29 In the present study, we found miR-381 was significantly downregulated in CRC with distant metastasis and TNM stage (Table 1), suggesting miR-381 might be a regulator of EMT. This was validated by our observation that miR-381 increased epithelial marker E-cadherin and inhibited mesenchymal markers N-cadherin and vimentin. Moreover, this phenomenon was dramatically attenuated by rescued Twist1 in miR-381 expressing HT29 and SW480 cells. These data suggest that miR-381 is a potential EMT inhibitor, at least partially, by targeting EMT inducer Twist1 in CRC cells.
Conclusion
In conclusion, our current findings revealed that miR-381 is downregulated in CRC and inhibits CRC invasion and migration by targeting Twist 1. These data suggest that restoration of miR-381 may be a potential therapeutic strategy for the patients with CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
Gill S, Thomas RR, Goldberg RM. Review article: colorectal cancer chemotherapy. Aliment Pharmacol Ther. 2003;18(7):683–692. | ||
Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. | ||
Geiger TR, Peeper DS. Metastasis mechanisms. Biochim Biophys Acta. 2009;1796(2):293–308. | ||
Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. | ||
Amirkhah R, Schmitz U, Linnebacher M, Wolkenhauer O, Farazmand A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosomes Cancer. 2015;54(3):129–141. | ||
Dassow H, Aigner A. MicroRNAs (miRNAs) in colorectal cancer: from aberrant expression towards therapy. Curr Pharm Des. 2013;19(7):1242–1252. | ||
Zhai H, Ju J. Implications of microRNAs in colorectal cancer development, diagnosis, prognosis, and therapeutics. Front Genet. 2011;2(78). | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. | ||
Xuan Y, Yang H, Zhao L, et al. MicroRNAs in colorectal cancer: small molecules with big functions. Cancer Lett. 2015;360(2):89–105. | ||
Feng Y, Zhu J, Ou C, et al. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer. 2014;110(9):2300–2309. | ||
Qian X, Yu J, Yin Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12(9):1385–1394. | ||
Kern HB, Niemeyer BF, Parrish JK, et al. Control of MicroRNA-21 expression in colorectal cancer cells by oncogenic epidermal growth factor/Ras signaling and Ets transcription factors. DNA Cell Biol. 2012;31(8):1403–1411. | ||
Shan L, Ji Q, Cheng G, et al. Diagnostic value of circulating miR-21 for colorectal cancer: a meta-analysis. Cancer Biomark. 2015;15(1):47–56. | ||
Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol Res Pract. 2015;211(8):557–569. | ||
Yan Q, Zhang W, Wu Y, et al. KLF8 promotes tumorigenesis, invasion and metastasis of colorectal cancer cells by transcriptional activation of FHL2. Oncotarget. 2015;6(28):25402–25417. | ||
de Krijger I, Mekenkamp LJ, Punt CJ, Nagtegaal ID. MicroRNAs in colorectal cancer metastasis. J Pathol. 2011;224(4):438–447. | ||
Hur K. MicroRNAs: promising biomarkers for diagnosis and therapeutic targets in human colorectal cancer metastasis. BMB Rep. 2015;48(4):217–222. | ||
Ma Y, Li W, Wang H. Roles of miRNA in the initiation and development of colorectal carcinoma. Curr Pharm Des. 2013;19(7):1253–1261. | ||
Ming J, Zhou Y, Du J, et al. MiR-381 suppresses C/EBPalpha-dependent Cx43 expression in breast cancer cells. Biosci Rep. 2015;35(6). | ||
Liang HQ, Wang RJ, Diao CF, Li JW, Su JL, Zhang S. The PTTG1-targeting miRNAs miR-329, miR-300, miR-381, and miR-655 inhibit pituitary tumor cell tumorigenesis and are involved in a p53/PTTG1 regulation feedback loop. Oncotarget. 2015;6(30):29413–29427. | ||
Formosa A, Markert EK, Lena AM, et al. MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c, miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p, mapped to the 14q32.31 locus, regulate proliferation, apoptosis, migration and invasion in metastatic prostate cancer cells. Oncogene. 2014;33(44):5173–5182. | ||
Liang Y, Zhao Q, Fan L, et al. Down-regulation of MicroRNA-381 promotes cell proliferation and invasion in colon cancer through up-regulation of LRH-1. Biomed Pharmacother. 2015;75:137–141. | ||
Martin A, Cano A. Tumorigenesis: Twist1 links EMT to self-renewal. Nat Cell Biol. 2010;12(10):924–925. | ||
Yang Z, Zhang X, Gang H, et al. Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem Biophys Res Commun. 2007;358(3):925–930. | ||
Tang H, Wang Z, Liu Q, Liu X, Wu M, Li G. Disturbing miR-182 and -381 inhibits BRD7 transcription and glioma growth by directly targeting LRRC4. PLoS One. 2014;9(1):e84146. | ||
Yan X, Yan L, Liu S, Shan Z, Tian Y, Jin Z. N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol Med Rep. 2015;12(2):2999–3006. | ||
Findlay VJ, Wang C, Watson DK, Camp ER. Epithelial-to-mesenchymal transition and the cancer stem cell phenotype: insights from cancer biology with therapeutic implications for colorectal cancer. Cancer Gene Ther. 2014;21(5):181–187. | ||
Pasquier J, Abu-Kaoud N, Al Thani H, Rafii A. Epithelial to mesenchymal transition in a clinical perspective. J Oncol. 2015;2015:792182. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.