Back to Journals » OncoTargets and Therapy » Volume 12
miR-3691-5p promotes hepatocellular carcinoma cell migration and invasion through activating PI3K/Akt signaling by targeting PTEN
Received 9 March 2019
Accepted for publication 16 May 2019
Published 21 June 2019 Volume 2019:12 Pages 4897—4906
DOI https://doi.org/10.2147/OTT.S208127
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Wei Du,1,* Xu Zhang,1,* Zhen Wan2
1Department of Clinical Medicine, School of Queen Mary, Nanchang University, Nanchang, 330006, People’s Republic of China; 2Department of General Surgery, the First Affiliated Hospital of Nanchang University, Nanchang 33006, People’s Republic of China
*These authors contributed equally to this work
Background: The enhanced ability of cancer metastasis is the major cause for the cancer-related death of hepatocellular carcinoma (HCC). Better understanding the mechanisms for the motility of cancer cells will benefit the treatment. Accumulating evidence suggests that aberrant microRNA (miRNA) expression contributes to HCC development and progression, whereas miR-3691-5p has not been reported in HCC.
Purpose: The aim of this study was to elucidate the expression, function and mechanism of miR-3691-5p in HCC.
Methods: Real-time quantitative polymerase chain reaction (qPCR) was performed to detect miR-3691-5p expression in HCC tissues and cell lines database analysis were conducted for detection of the expression of miR-3691-5p in HCC. Then, the association of miR-3691-5p with clinicopathological features of HCC patients were statistically measured. Subsequently, we attempted to observe the effects of miR-3691-5p on migration and invasion of HCC cells by transwell assays. Furthermore, bioinformatics tools and luciferase reporter gene assay as well as recuse experiments were conducted to explore the target of miR-3691-5p in HCC, and to explore whether the target mediated the effects of miR-3691-5p HCC cells.
Results: In the current study, we found that miR-3691-5p expression was elevated in both HCC tissues and cell lines, which was significantly correlated with poor prognosis and clinicopathological features including TNM stage (P=0.016) and vascular invasion (P=0.016). Furthermore, gain-or loss-of function assays demonstrated that miR-3691-5p promoted HCC cell migration and invasion. Luciferase reporter assay confirmed that PTEN was a direct downstream target of miR-3691-5p. Recuse assays showed that restoration of PTEN reversed the effects of miR-3691-5p on HCC cell migration and invasion through decreasing PI3K/Akt signaling.
Conclusion: Our results demonstrated that miR-3691-5p contributes to HCC cell migration and invasion through activating PI3K/Akt signaling by targeting PTEN.
Keywords: miR-3691-5p, hepatocellular carcinoma, PTEN, PI3K/Akt, migration, invasion
Introduction
Hepatocellular carcinoma (HCC), one of the most common cancer, represents the third leading cause of cancer-related mortality worldwide.1–3 China alone accounts for about half of the total number of cases and deaths due to the prevalence of hepatitis B virus.4,5 In spite of advances in diagnostic and treatment strategies, the prognosis of patients with HCC remains poor because of the metastasis and recurrence.6,7 For these reasons, the identification of key molecules involved in HCC progression is urgent and highly demanded for improving the clinical outcome.
microRNAs (miRNAs) are small non-coding RNA molecules which regulate target genes by a post-transcriptional mechanism. Extensive studies indicate that miRNAs function as tumor promoting or suppressive factors to regulate tumor cell phenotypes including cell proliferation,8 cell cycle,9 invasion,10,11 migration,12,13 autophagy,14 and cellular senescence.15 However, whether and how miR-3691-5p is involved in the progress of HCC metastasis, invasion and migration is unknown.
Phosphatase and tensin homolog (PTEN) as a tumour suppressor has been extensively studied in several tumor types including glioblastoma,16 breast cancer,17 endometrial cancer,18 ovarian cancer,19 lung cancer,20 prostate cancer,21 colorectal cancer22 and HCC.23 Previous studies have indicated a plethora of different miRNAs have been linked to PTEN regulation in human cancers including miR-19a in leukaemia,24 miR-22 in prostate25 and miR-26a in high-grade glioma26 and miR-21 in HCC.27 However, the relationship between PTEN and miR-3691-5p in HCC remains unknown.
In the present study, we demonstrated a significant increase of miR-3691-5p in HCC tissues and cells, which was correlated with poor prognosis and clinic pathological features. Our studies showed that miR-3691-5p promoted HCC cell migration and invasion through activating PI3K/Akt signaling by targeting PTEN. In conclusion, we demonstrated that miR-3691-5p functions as an oncogene in HCC.
Methods and materials
Tissue samples
Tissue samples were obtained from 43 patients who were undergoing liver resection in the Department of General Surgery at the First Affiliated Hospital of Nanchang University (Nanchang, China). The HCC patients did not receive any adjuvant therapy before surgery, such as chemotherapy or radiotherapy. All HCC and normal tissues were collected and restored in liquid nitrogen. All the patients were provided written informed consent. Approval was obtained from the Ethics Committee of Nanchang University. The clinicopathologic parameters of patients were shown in Table 1.
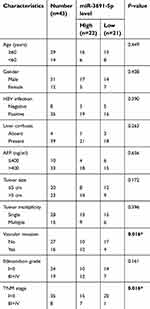 | Table 1 Association between miR-3691-5p expression and clinicopathologic features of patients with hepatocellular carcinoma |
Cell culture, transfection and reagent
HEK 293T, the human HCC cell lines (Hep3B, SMMC-7721, MHCC97-L, MHCC97-H, HCCLM3) and the human immortalized normal hepatic cell line L02 were obtained from the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS; Gibco) with 1% penicillin-streptomycin (Sigma, St-Louis, MO, USA) in a humidified incubator containing of 5% CO2 at 37 °C.
The mimics control (miR-control, CmiR-SN0001-SN), miR-3691-5p mimics (miR-3691-5p, HmiR-SN1977-SN-12), inhibitors control (anti-miR-NC, CmiR-AN0001-SN) and miR-3691-5p inhibitors (anti-miR-3691-5p, HmiR-AN1977-SN-20) were purchased from Genecopoeia (Guangzhou, China). PTEN expression plasmid (PTEN, SC119965), specific siRNA against PTEN (si-PTEN, SR321496) and their corresponding negative control (EV and si-control, PS100001 and SR30004) were constructed and purchased from OriGene (OriGene Technologies Inc., Rockville, MD, USA), and the sequences for PTEN siRNA and scrambled siRNA were shown in Table 2. Cells transfection was performed with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. An Akt inhibitor, MK2206 (1 μM; Selleck Chemicals, Houston, TX, USA) was used to treat HCC cells for 12 h.
 | Table 2 Primers used in this study |
Real-time quantitative polymerase chain reaction (qPCR)
Trizol (Invitrogen) was used to extract RNA from tissues and cells following as the manufacture procedure. TIANScript RT Kit (Tiangen Biotech, Beijing, China) was used to perform reverse transcription. Quantitative PCR were performed using TaqMan Human MiRNA Aaasy Kit (Genecopoeia) and SYBR Premix Ex TaqTM Kit (TaKaRa, Shiga, Japan). The primers for PTEN (HQP067630) and GAPDH (HQP006940) were purchased from Genecopoeia. The primers for miR-3691-5p (HmiRQP1977) and U6 (HmiRQP9001) were obtained from Genecopoeia. The sequences of primers have been shown in Table 3. Expression levels were quantified using the 2−ΔΔCt method. Each experiment was performed in triplicate.
 | Table 3 Primers used in this study |
Cell migration and invasion assays
Transwell chambers (Millipore, Burlington, MA, USA) were used for cell migration and invasion assays. For migration assays, HCC cells (2×104) in the upper chamber were cultured with serum-free DMEM, and the lower chamber was filled with 10% serum-containing DMEM. For invasion assays, HCC cells (2×104) were seeded on Matrigel-coated membrane inserts. Then, the chamber was put into the cell culture plate and incubated at 37 °C for 24 hrs. Subsequently, cells that migrated or invaded across the transwell membrane were fixed with 4% paraformaldehyde for 10 min and next stained with 0.1% crystal violet for 20 min. Finally, the migratory and invasive cells were examined and counted under microscope.
Western blotting
Proteins from HCC cells and tissues were extracted with RIPA buffer, the concentration was measured with the BCA Kit. Then proteins were electrophoresed by 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad, Hercules, CA, USA). Next, the membranes were blocked in 5% non-fat milk and incubated with specific primary antibodies at 4 °C overnight. PTEN (1:1000, #9188; Cell Signaling Technology, Danvers, MA, USA), Akt (1:1000, #4691; Cell Signaling Technology), p-Akt (1:2000, #4060; Cell Signaling Technology), GAPDH (1:1000, #5174; Cell Signaling Technology) antibodies were used. After that, the membranes were incubated with the second antibody (anti‑rabbit #7074; Cell Signaling Technology) for 1 h at room temperature. Finally, the ECL reagent (Beyotime Institute of Biotechnology, Haimen, China) was applied for detection.
Luciferase reporter assay
The wild-type (WT) or mutant (MT) 3’UTR of PTEN mRNA were synthesized and inserted into downstream of the pEZX-MT06 vector (Genecopoeia). HEK 293T cells transfected with miR-3691-5p mimics or inhibitors or the corresponding controls were transfected with PTEN −3’UTR-wt and PTEN −3’UTR-mut, respectively. Cells were collected 48 h later and the luciferase activity was quantified using the Luc-PairTM Duo- Luciferase Assay Kit (Genecopoeia).
Statistical analysis
Data are presented as the mean ± standard and analyzed using GraphPad Prism 7.0 software. All experiments were repeated for at least 3 times. Student’s t-test (two groups) and one-way ANOVA (multiple groups) were conducted to analyze the difference. Spearman’s correlation analysis was performed to determine the correlation between miR-3691-5p and PTEN mRNA expression. *P<0.05, **P<0.01 and ***P<0.001 were taken as indicative of statistically significant difference.
Results
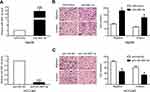
Expression and clinical significance of miR-3691-5p in HCC
Real-time quantitative PCR (qPCR) was performed to detect miR-3691-5p expression in 43 pairs HCC tissues and the adjacent non‑tumor tissues. The results showed a higher expression of miR-3691-5p in HCC tissues than in non‑tumor tissues (P<0.001, Figure 1A). In accordance, higher miR-3691-5p level was observed in HCC cell lines (Hep3B, SMMC-7721, MHCC97-L, MHCC97-H, HCCLM3) compared to the immortalized normal liver cell line (L02) (Figure 1C). TCGA data from starBase v3.0 (
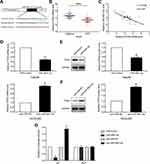
miR-3691-5p promotes HCC cell migration and invasion
To investigate the biological function of miR-3691-5p on the migration and invasion of HCC cells in vitro, we constructed both miR-3691-5p overexpression and knockdown HCC cell lines using miR-3691-5p mimics in Hep3B and inhibitors in HCCLM3 cells, respectively. Transfection efficiency in Hep3B or HCCLM3 cells was confirmed by qPCR (Figure 2A). Results of transwell assay with or without matrigel showed that miR-3691-5p overexpression notably enhanced Hep3B cell migration and invasion, while knockdown of miR-3691-5p significantly inhibited HCCLM3 cell migration and invasion compared with controls (Figure 2B and C). These data suggested that miR-3691-5p promotes HCC cell migration and invasion.
PTEN is a direct downstream of miR-3691-5p in HCC
TargetScan (
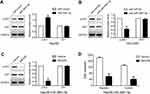
PTEN mediates the role of miR-3691-5p in HCC cells
Rescue assays were conducted to illuminate whether PTEN participates in the facilitation of miR-3691-5p on migration and invasion in HCC cells. We increased PTEN expression in miR-3691-5p-overexpressing Hep3B cells (Figure 4A), and results of transwell assays revealed that the abilities of migration and invasion were weakened after PTEN upregulation (P<0.05, Figure 4B). Accordingly, PTEN silence with siRNAs enhanced the abilities of migration and invasion of HCCLM3-anti-miR-3691-5p cells (P<0.05, Figure 4A and C). These data showed that PTEN is a functional downstream mediator of miR-3691-5p in HCC.
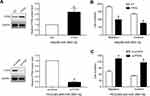
PI3K/Akt pathway is essential for the biological function of miR-3691-5p in HCC
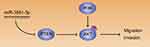
Given that PTEN was a major negative regulator of PI3K/Akt pathway, we further investigated whether miR-3691-5p could altered the activity of PI3K/Akt pathway in HCC cells. Results of Western blotting revealed that miR-3691-5p mimics increased the expression of p-Akt in Hep3B cell and miR-3691-5p inhibitors decreased the expression of p-Akt in HCCLM3 (P<0.05, Figure 5A and B). To investigate whether PI3K/Akt pathway mediates the effects of miR-3691-5p on migration and invasion of HCC cells, MK2206 (an Akt inhibitor) was used to block Akt activation in Hep3B cells with miR-3691-5p overexpression (P<0.05, Figure 5C). Functional experiments revealed that inactivation of Akt inhibited miR-3691-5p-overexpressing Hep3B cells migration and invasion (P<0.05, Figure 5D). These results demonstrated that PI3K/ Akt signaling exerts an essential function during miR-3691-5p-induced HCC cell migration and invasion (Figure 6).
Discussion
In the recent years, increasing evidence has demonstrated that miRNAs, as non-coding RNAs, plays a vital role in the regulation of HCC progression.28–30 For example, dysregulation of miRNAs promotes the growth31,32 and metastasis33,34 of HCC cells. Hence, identification of HCC specific miRNAs and their targets are critical for understanding their role in HCC progression and may be important for defining novel therapeutic targets. To our knowledge, the study is the first time to investigate the relationship between miR-3691-5p and HCC. In this study, we revealed that miR-3691-5p was upregulated in HCC tissues and cell lines, which was significantly correlated with poor prognosis and clinic pathological features including TNM stage and vascular invasion. Our results demonstrated that miR-3691-5p promoted HCC cell migration and invasion by gain and loss of function experiments. In conclusion, we demonstrated that miR-3691-5p functions as an oncogene in HCC.
Ample evidence has shown that a variety of different miRNAs have been linked to PTEN regulation in human cancers including miR-19a in leukaemia,24 miR-22 in prostate25 and miR-26a in high-grade glioma26 and miR-21 in HCC.27 However, the relationship between PTEN and miR-3691-5p has not been investigated in HCC. Here, we provided sufficient evidence that PTEN is a direct function target of miR-3691-5p in HCC. First, the level of miR-3691-5p was inversely correlated with the expression of PTEN in HCC tissues. Second, miR-3691-5p negatively modulated PTEN expression in HCC cells at the mRNA and protein levels. Third, miR-3691-5p affected the luciferase activity of wt-3’UTR but not of the mut-3’UTR of PTEN. Finally, recuse assays showed that restoration of PTEN reverses the effects of miR-3691-5p on HCC cell migration and invasion through decreasing PI3K/Akt signaling. These results suggested that PTEN is a direct function target of miR-3691-5p, and mediates the role of miR-3691-5p in HCC cells.
In this study, we found that miR-3691-5p was significantly elevated in HCC tissues and cell lines, and correlated with poor prognosis and clinic pathological features, which promotes HCC cell migration and invasion through activating PI3K/Akt signaling by targeting PTEN. In conclusion, our results demonstrated that miR-3691-5p functions as an oncogene in HCC
Acknowledgments
This work was supported by the grants from National Nature Science Foundation of China (No. 81760137), Jiangxi Province Science Foundation for Youths (No. 20161BAB215225) and Science and Technology Project Founded by the Education Department of Jiangxi Province (No. 150131).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
2. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi:10.3322/caac.21349
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi:10.3322/caac.21442
4. Islami F, Chen W, Yu XQ, et al. Cancer deaths and cases attributable to lifestyle factors and infections in China, 2013. Ann Oncol. 2017;28:2567–2574. doi:10.1093/annonc/mdx342
5. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
6. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi:10.1016/S0140-6736(18)30010-2
7. Greten TF, Wang XW, Korangy F. Current concepts of immune based treatments for patients with HCC: from basic science to novel treatment approaches. Gut. 2015;64:842–848. doi:10.1136/gutjnl-2014-307990
8. Dou C, Zhou Z, Xu Q, et al. Hypoxia-induced TUFT1 promotes the growth and metastasis of hepatocellular carcinoma by activating the Ca(2+)/PI3K/AKT pathway. Oncogene. 2019;38:1239–1255. doi:10.1038/s41388-018-0505-8
9. Ma YS, Lv Z-W, Yu F, et al. MicroRNA-302a/d inhibits the self-renewal capability and cell cycle entry of liver cancer stem cells by targeting the E2F7/AKT axis. J Exp Clin Cancer Res. 2018;37:252. doi:10.1186/s13046-018-0927-8
10. Luo C, Yin D, Zhan H, et al. microRNA-501-3p suppresses metastasis and progression of hepatocellular carcinoma through targeting LIN7A. Cell Death Dis. 2018;9:535. doi:10.1038/s41419-018-0577-y
11. Liu Z, Wang Y, Dou C, et al. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49. doi:10.1186/s13046-018-0717-3
12. Roy S, Hooiveld GJ, Seehawer M, et al. microRNA 193a-5p regulates levels of nucleolar- and spindle-associated protein 1 to suppress hepatocarcinogenesis. Gastroenterology. 2018;155:1951–1966 e1926. doi:10.1053/j.gastro.2018.08.032
13. Tao J, Liu Z, Wang Y, et al. MicroRNA-645 represses hepatocellular carcinoma progression by inhibiting SOX30-mediated p53 transcriptional activation. Int J Biol Macromol. 2019;121:214–222. doi:10.1016/j.ijbiomac.2018.10.032
14. Wang J, Chen J, Liu Y, et al. Hepatitis B virus induces autophagy to promote its replication by the Axis of miR-192-3p-XIAP via NF-kappaB signaling. Hepatology. 2018. doi:10.1002/hep.30248
15. Shiu TY, Shih Y-L, Feng A-C, et al. HCV core inhibits hepatocellular carcinoma cell replicative senescence through downregulating microRNA-138 expression. J Mol Med. 2017;95:629–639. doi:10.1007/s00109-017-1518-4
16. Benitez JA, Ma J, D’Antonio M, et al. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat Commun. 2017;8:15223. doi:10.1038/ncomms15223
17. Ngeow J, Sesock K, Eng C. Breast cancer risk and clinical implications for germline PTEN mutation carriers. Breast Cancer Res Treat. 2017;165:1–8. doi:10.1007/s10549-015-3665-z
18. Bian X, Gao J, Luo F, et al. PTEN deficiency sensitizes endometrioid endometrial cancer to compound PARP-PI3K inhibition but not PARP inhibition as monotherapy. Oncogene. 2018;37:341–351. doi:10.1038/onc.2017.326
19. Wang Y, Zhao S, Zhu L, Zhang Q, Ren Y. MiR-19a negatively regulated the expression of PTEN and promoted the growth of ovarian cancer cells. Gene. 2018;670:166–173. doi:10.1016/j.gene.2018.05.063
20. Malaney P, Palumbo E, Semidey-Hurtado J, et al. PTEN physically interacts with and regulates E2F1-mediated transcription in lung cancer. Cell Cycle. 2018;17:947–962. doi:10.1080/15384101.2017.1388970
21. Wise HM, Hermida MA, Leslie NR. Prostate cancer, PI3K, PTEN and prognosis. Clin Sci. 2017;131:197–210. doi:10.1042/CS20160026
22. Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852–864. doi:10.3109/0284186X.2014.895036
23. Wan XW, Jiang M, Cao H-F, et al. The alteration of PTEN tumor suppressor expression and its association with the histopathological features of human primary hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129:100–106. doi:10.1007/s00432-002-0410-x
24. Li Y, Vecchiarelli-Federico LM, Li Y-J, et al. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119:4486–4498. doi:10.1182/blood-2011-09-378687
25. Pasqualini L, Bu H, Puhr M, et al. miR-22 and miR-29a are members of the androgen receptor cistrome modulating LAMC1 and Mcl-1 in prostate cancer. J Mol Endocrinol. 2015;29:1037–1054. doi:10.1210/me.2014-1358
26. Huse JT, Brennan C, Hambardzumyan D, et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi:10.1101/gad.1777409
27. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi:10.1053/j.gastro.2007.05.022
28. Wu H, Zhang W, Wu Z, et al. miR-29c-3p regulates DNMT3B and LATS1 methylation to inhibit tumor progression in hepatocellular carcinoma. Cell Death Dis. 2019;10:48. doi:10.1038/s41419-018-1281-7
29. Valdmanis PN, Kim HK, Chu K, et al. miR-122 removal in the liver activates imprinted microRNAs and enables more effective microRNA-mediated gene repression. Nat Commun. 2018;9:5321. doi:10.1038/s41467-018-07786-7
30. Teufel M, Seidel H, Köchert K, et al. Biomarkers associated with response to regorafenib in patients with hepatocellular carcinoma. Gastroenterology. 2019;156:1731–1741. doi:10.1053/j.gastro.2019.01.261
31. Liao ZB, Tan XL, Dong KS, et al. miRNA-448 inhibits cell growth by targeting BCL-2 in hepatocellular carcinoma. Dig Liver Dis. 2018. doi:10.1016/j.dld.2018.09.021
32. Bao J, Yu Y, Chen J, et al. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis. 2018;9:1045. doi:10.1038/s41419-018-1020-0
33. Shi D-M, Li L-X, Bian X-Y, et al. miR-296-5p suppresses EMT of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3 signaling. J Exp Clin Cancer Res. 2018;37:294. doi:10.1186/s13046-018-0957-2
34. Liu Z, Li W, Pang Y, et al. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68. doi:10.1016/j.ebiom.2018.10.067
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.