Back to Journals » OncoTargets and Therapy » Volume 9
miR-34b inhibits nasopharyngeal carcinoma cell proliferation by targeting ubiquitin-specific peptidase 22
Authors Xiao J, Li Y, Zhang W, Jiang Y, Du B, Tan Y
Received 15 October 2015
Accepted for publication 28 January 2016
Published 16 March 2016 Volume 2016:9 Pages 1525—1534
DOI https://doi.org/10.2147/OTT.S98378
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Faris Farassati
Jianyong Xiao,1 Yingying Li,2 Wenyin Zhang,1 Yanni Jiang,1 Biaoyan Du,3 Yuhui Tan1
1Department of Biochemistry, Guangzhou University of Chinese Medicine, 2Department of Internal Medicine, Guangzhou Eighth People’s Hospital, 3Department of Pathology, Guangzhou University of Chinese Medicine, Guangzhou, People’s Republic of China
Objectives: This study aimed to investigate the precise role of miR-34b in nasopharyngeal carcinoma (NPC).
Materials and methods: The expression of miR-34b and transcription of ubiquitin-specific peptidase 22 (USP22) were examined using quantitative reverse transcription-polymerase chain reaction. Western blot analysis was used to measure the protein expression of USP22. A dual-luciferase assay was used to investigate the interaction between miR-34b and USP22. Cell viability was determined by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay. The cell cycle was analyzed by propidium iodide staining followed by flow cytometry analysis.
Results: miR-34b was significantly downregulated in NPC tissues and NPC cell lines. Overexpression of miR-34b in NPC SUNE-6-10B cells inhibited cell viability and proliferation. USP22 was highly expressed in NPC cells and promoted cell viability and proliferation. Restoration of USP22 expression could reverse the effect of miR-34b on NPC cell viability and proliferation.
Conclusion: miR-34b acts as a tumor suppressor in NPC, which is mediated via repression of the oncogene USP22.
Keywords: cell proliferation, miR-34b, nasopharyngeal carcinoma, overexpression, USP22
Introduction
Nasopharyngeal carcinoma (NPC) derived from the nasopharyngeal epithelium has a high incidence rate in Southeast Asia and Northern Africa.1 Genetic susceptibility, endemic environmental factors, and Epstein–Barr virus infection are believed to be the major etiological factors of NPC.2 Currently, radiotherapy is the most sensitive and effective treatment for NPC, but the prognosis is often not satisfactory due to the rates of recurrence and metastasis.3 Elucidation of the molecular mechanisms underlying its pathogenesis and progression is essential for the development of novel strategies for the diagnosis and treatment of NPC.
MicroRNAs (miRNAs) are noncoding small RNAs that can regulate their target gene expression to modulate cell proliferation, migration, differentiation, and apoptosis.4 Abnormal miRNAs can function as either oncogenes or tumor suppressors.5 Multiple miRNAs have been shown to be aberrantly expressed and to contribute to the development and progression of NPC.6 Although miR-34b has been reported to be a tumor suppressor gene in many types of cancers, its function in NPC remains not well understood. The miRNA expression profile showing that miR-34b is underexpressed in NPC compared to normal tissue samples caught our attention.7,8 In a recent study, we compared the expression of miR-34b in NPC cell lines and an immortalized nasopharyngeal epithelial cell line and found that miR-34b was also downregulated in NPC cells. Furthermore, we performed a bioinformatics analysis of the target genes of miR-34b and selected ubiquitin-specific peptidase 22 (USP22) as a gene of interest.
USP22, a newly discovered member of the ubiquitin hydrolase family, possesses deubiquitinating activity.9 USP22 expression is significantly elevated in tumor tissues, compared with normal tissues,10 and it has been suggested as a valuable biomarker for predicting the recurrence and metastasis of malignancies.11,12 USP22 plays a key role in cell cycle regulation, and interference with USP22 expression in tumor cells can arrest cell cycle progression in the G1 phase and inhibit tumor cell proliferation.13,14 USP22 was also identified as a subunit of the Spt–Ada–Gcn5 acetyltransferase coactivator complex, which regulates the expression of genes related to oncogenicity and proliferation.15 However, little is known about the oncogenic role played by USP22 in the growth of NPC.
In this study, we evaluated the impact of miR-34b overexpression on the proliferation of NPC. Furthermore, we identified the oncogene USP22 as a target gene of miR-34b. Overexpression of USP22 attenuates the inhibitory effect of miR-34b on NPC cell viability and proliferation, thus confirming our hypothesis that miR-34b inhibits NPC proliferation by downregulating USP22.
Materials and methods
Cell lines
All human/animal studies have been approved by the Institute Research Medical Ethics Committee of Guangzhou University of Chinese Medicine. All human studies have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. All persons gave their informed verbal consent prior to their inclusion in the study. Human NPC cell lines CNE-1, CNE-2, SUNE-5-8F, and SUNE-6-10B were preserved in our laboratory and maintained in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C in a humidified atmosphere containing 5% CO2. The control cell line NP69 was purchased from the Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences and maintained in Keratinocyte Serum-Free Medium (Thermo Fisher Scientific) in a humidified atmosphere (37°C, 5% CO2). When the cells reached the logarithmic growth phase, the subsequent experimental analyses were performed.
RNA isolation and quantitative reverse transcription-polymerase chain reaction
Total RNA was isolated from the cell lines and tissue samples using TRIzol (Thermo Fisher Scientific), according to the manufacturer’s instructions. mRNA was reverse transcribed using the ReverTra Ace qPCR RT Kit (Toyobo Biochemicals, Toyko, Japan), according to the manufacturer’s protocol. Real-time polymerase chain reaction (PCR) was performed with GoTaq® qPCR Master Mix (Promega Corporation, Fitchburg, WI, USA) using a MiniOpticon™ Real-Time PCR detection instrument (Bio-Rad, Hercules, CA, USA) with the SyBr Green detection protocol provided by the manufacturer. Reverse transcription and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) for miRNA were performed using the Stem-Loop miRNA qRT-PCR Primer Set (Forevergen, Guangzhou, People’s Republic of China). Data analysis was performed using the 2−ΔΔCt method. The level of USP22 was normalized to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The level of miR-34b was normalized to the expression of U6. Sequence-specific primers for USP22 and GAPDH were as follows: USP22, 5′-CCATTGATCTGATGTACGGAGG-3′ (forward) and 5′-TCCTTGGCGATTATTTCCATGTC-3′ (reverse) and GAPDH, 5′-GAGTCAACGGATTTGGTCGT-3′ (forward) and 5′-GACAAGCTTCCCGTTCTCAG-3′ (reverse). The primers used for miR-34b and U6 were purchased from Forevergen.
Western blot analysis
Cells from all the experimental groups were collected using a cell scraper, and proteins were extracted using cell lysis buffer. The protein concentration was assessed by Bradford assay, and equal amounts of total protein were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and blocked with 5% skim milk for 1 hour at room temperature. The polyvinylidene fluoride membranes were then washed with TBST (containing NaCl, Tris–HCl, and Tween-20) and incubated overnight with primary antibodies against target proteins at 4°C followed by two washes in TBST. The USP22 antibody was purchased from Abcam (Abcam, Cambridge, MA, USA). The membranes were incubated with the appropriate secondary antibodies for 1 hour at room temperature and then washed three times with TBST. Proteins were visualized by chemiluminescence (Forevergen).
Plasmid construction and transfection
The 3′-untranslated region (3′-UTR) of USP22 genes was cloned into the pmirGlo plasmid (Promega Corporation) between the SacI and XbaI sites using the following primers: USP22 wild-type 3′-UTR forward primer, 5′-ATGCGAGCTCTAGACAGCCAGGGAGTAAACAC-3′ and reverse primer, 5′-ATGCTCTAGATGACATAGGAGAGACTACAAAGCA-3′. The pmirGlo plasmids containing the mutated USP22 3′-UTR were constructed using a KOD-plus Mutagenesis Kit (Toyobo Biochemicals) according to the manufacturer’s instructions. Primers for mutated USP22 3′-UTR were as follow: 5′-CAAAACTTACTGATAAATTGTCAAAAGAACA-3′ (forward) and 5′-AATTTATCAGTAAGTTTTGTTTACTGTAAGTTTG-3′ (reverse). The open reading frame expression vectors pCMV-USP22 and the control vector pCMV were purchased from the GeneChem Corporation (GeneChem Corporation, Shanghai, People’s Republic of China). All the inserted or mutated sequences were confirmed by sequencing. All the transfection reactions were performed using Lipofectamine 2000 (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Transfection of miRNAs and small interfering siRNAs
All the small interfering RNAs (siRNAs) and miRNA mimics were purchased from Guangzhou RiboBio (RiboBio, Guangzhou, People’s Republic of China) and transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific), as recommended by the manufacturer. The cells were transfected with miRNA mimics or siRNA at a concentration of 100 nM. The sequences of the siRNAs were as follows: siRNA-USP22: sense, 5′-CACGGACAGTCTCAACAAT-3′ and antisense, 5′-ATTGTTGAGACTGTCCGTG-3′.
3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay
Harvested cells were seeded in 96-well plates at 5×103 cells/well and grown in normal medium for 1 day, 2 days, 3 days, and 4 days. Subsequently, the cells were incubated in 0.1 mg/mL 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) at 37°C for 3 hours and lysed in dimethyl sulfoxide at room temperature for 30 minutes. The absorbance in each well was measured at 490 nm using a microplate reader. Each experiment was performed in triplicate.
Colony forming assay
Colony forming assays were performed as described previously.16 Cells were seeded into triplicate six-well plates at a density of 1,000 cells/well, and the medium was replaced every 3 days. After 10 days, the cells were washed twice with phosphate-buffered saline (PBS), fixed, and stained with 0.5% crystal violet. The colonies were then counted under a microscope.
Cell cycle analysis by flow cytometry
Cells were trypsinized, collected, and washed three times with PBS. The cells were then fixed in 1 mL of 70% ice-cold ethanol overnight at 4°C. After two washes with PBS and centrifugation for 10 minutes at 1,000 rpm, the supernatant was discarded and the cell pellets were stained with 50 μg/mL of propidium iodide and 100 U/mL of RNase A and incubated in PBS for 30 minutes at room temperature in dark conditions. The percentages of cells in different phases of the cell cycle were determined by evaluating the DNA content. A total of 10,000 cells/sample were counted. A cytometric analysis was performed with a flow cytometer (BD Biosciences, San Jose, CA, USA) using CellQuest software. Each assay was performed in triplicate.
Dual-luciferase assay
Cells were seeded in 48-well plates 1 day prior to transfection. The miRNA mimics (100 nM) and plasmid (5 ng/mL) were cotransfected into SUNE-6-10B cells. At 48 hours after transfection, the luciferase activity was measured using the Dual-Glo luciferase assay kit (Promega Corporation), according to the manufacturer’s instructions.
Statistics analysis
SPSS 17.0 (SPSS Inc., Chicago, IL, USA) was used to analyze the experimental data. All the experiments were performed in triplicate, and the results were analyzed by analysis of variance or Student’s t-test and are expressed as mean ± SD. P-value <0.05 or <0.01 was considered to be statistically significant.
Results
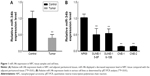
miR-34b is downregulated in NPC tissue samples and cell lines
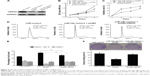
To investigate the role of miR-34b in NPC, we first compared its expression levels in NPC and adjacent noncancerous tissue samples (sample size: tumor group: n=18 and control group: n=15). NPC samples showed significant downregulation of miR-34b compared with adjacent noncancerous tissue samples (Figure 1A). We further compared the expression levels of miR-34b in NPC cell lines (SUNE-5-8F, SUNE-6-10B, CNE-1, and CNE-2) against expression in the immortalized nasopharyngeal epithelial cell line NP69. The qRT-PCR analysis showed that the basal expression levels of miR-34b were generally downregulated in the four NPC cell lines compared with NP69 (Figure 1B). The downregulation of miR-34b in NPC cell lines suggests that miR-34b might be involved in the progression of NPC.
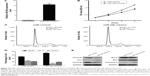
Overexpression of miR-34b inhibits cellular proliferation of NPC
To evaluate whether miR-34b has an effect on NPC cell proliferation, we transfected the NPC cell line SUNE-6-10B with miR-34b mimics and negative control (NC) mimics. The qRT-PCR analysis revealed a significant upregulation of miR-34b after transfection (Figure 2A). The MTT assay showed that cell viability was significantly suppressed in miR-34b-transfected cells in comparison with NC-transfected cells (Figure 2B). Cell cycle analysis demonstrated that miR-34b overexpression led to an increased percentage of SUNE-6-10B cells in the G1 phase and a decreased percentage of cells in the S phase, which suggests that miR-34b can induce G1 arrest (Figure 2C and D). To further confirm that miR-34b has an effect on NPC cell proliferation, expression levels of cell cycle-related proteins P21, CyclinD and CyclinE, and apoptosis-related proteins Bcl-2, Bax, and Caspase-3 were analyzed (Figure 2E). P21 was increased, while CyclinD and CyclinE were suppressed in the miR-34b mimics group. In addition, Bax and Caspase were increased and Bcl-2 was decreased in the miR-34b mimics group. These results indicate that miR-34b suppresses the proliferation of NPC cells.
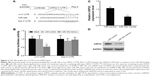
miR-34b targets USP22 to cause its downregulation
In order to gain insight into the mechanism by which miR-34b inhibits NPC, we searched for candidate target genes of miR-34b using bioinformatics and ultimately focused on USP22. TargetScan analysis revealed that the 3′-UTR sequence of USP22 contains one conserved miR-34b binding site (Figure 3A). To confirm that USP22 is a direct target of miR-34b, we cloned the 3′-UTR of USP22 into a reporter plasmid downstream of the luciferase gene. The reporter assays showed that the upregulation of miR-34b significantly decreased the relative luciferase activity of the wild-type 3′-UTR of USP22, compared with the NC, but had no effect on the mutant USP22 3′-UTR. These results suggest that miR-34b directly binds to the 3′-UTR of USP22 (Figure 3B). Next, we detected the expression of USP22 in SUNE-6-10B cells following transfection with miR-34b mimics and NC mimics. qRT-PCR and Western blot data show that miR-34b transfection significantly suppressed the level of USP22 (Figure 3C and D). These results suggest that USP22 is a direct downstream target gene of miR-34b.
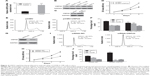
USP22 promotes the proliferation of NPC
qRT-PCR analysis showed that the expression levels of USP22 mRNA were higher in NPC tissues than that in normal nasopharyngeal mucosa (Figure 4A) (sample size: tumor group: n=18 and control group: n=15). We also examined the expression of USP22 in four NPC cell lines and NP69 cells. Western blot analysis showed that USP22 was highly expressed in four NPC cell lines compared to NP69 cells and exhibited an inverse expression pattern compared with miR-34b (Figure 4B). To explore the function of USP22 in NPC cells, we constructed the USP22 overexpression vector pCMV-USP22 (Figure 4C). The MTT assay showed that overexpression of USP22 increased the viability of SUNE-6-10B cells compared with pCMV (Figure 4D). The cell cycle distribution revealed that a high level of USP22 resulted in a decrease of SUNE-6-10B cells in the G1 phase and an increase of SUNE-6-10B cells in the S phase (Figure 4E and F).
Additionally, we transfected SUNE-6-10B cells with an siRNA against USP22 and observed that USP22 knockdown resulted in an impaired viability of SUNE-6-10B cells compared with the NC (Figure 4G). The MTT assay demonstrated that USP22 knockdown decreased the viability of SUNE-6-10B cells (Figure 4H). Flow cytometry revealed that USP22 siRNA transfection led to an increased percentage of SUNE-6-10B cells in the G1 phase and a decreased proportion of SUNE-6-10B cells in the S phase compared with the siRNA NC (Figure 4I and J).
These results demonstrate that USP22 promotes cell proliferation and plays an oncogenic role in NPC cells.
Overexpression of USP22 attenuates the inhibitory effect of miR-34b on NPC cell proliferation
To investigate whether miR-34b inhibits NPC cell proliferation by targeting USP22, we transfected SUNE-6-10B cells and a cell line that was stably expressing USP22 (Figure 5A) with miR-34b mimics. MTT (Figure 5B and C), cell cycle (Figure 5D and E), and colony formation analyses (Figure 5F) show that overexpression of USP22 can significantly attenuate the suppressive effect of miR-34b. These results provide further support that USP22 is a downstream functional target of miR-34b.
Discussion
NPC has a high incidence in southern People’s Republic of China and is difficult to detect during its early stages due to the lack of symptoms.17 At later stages, the reduced radiosensitivity and high invasiveness of NPC typically cause poor clinical outcomes.18 The development of more sensitive diagnostic markers and therapeutic targets are urgently needed. miRNAs, which are abundant in NPC,19 are ideal candidates because they are differentially expressed in NPC and normal nasopharyngeal tissues.20 Aberrant expression of miRNAs may represent a novel marker in the clinical diagnosis and prognosis of NPC.21 Importantly, miRNAs can also play tumor suppressor or oncogenic roles in NPC cells through the downregulation of their downstream target genes.16,22–25 In the present study, we evaluated the oncogene USP22 as a potential target gene of miR-34b. Furthermore, we demonstrated that miR-34b bound directly to the 3′-UTR of USP22 and downregulated its protein expression. The results of this study provide a novel insight of the mechanism by which miR-34b suppresses the proliferation of NPC cells.
Although miR-34b has been identified as a tumor suppressor in many types of cancers, it has also been reported to play an oncogenic role in breast cancer and esophageal squamous cell carcinoma.26,27 Therefore, we sought to evaluate the precise effect of miR-34b in NPC. To achieve this goal, we overexpressed miR-34b by transfection into NPC SUNE-6-10B cells and observed an inhibition of cell viability. Furthermore, flow cytometry analysis revealed an arrest of SUNE-6-10B cells in the G1 phase, which suggests that miR-34b functions as a negative regulator of cellular proliferation in NPC. Moreover, a comparison of the expression levels of miR-34b in NPC cell lines and immortalized nasopharyngeal epithelial cells revealed that the basal expression level of miR-34b was generally downregulated in four NPC cell lines compared with NP69 (Figure 1A). These results are consistent with miRNA expression profiling, which shows a downregulation of miR-34b in eight NPC tissues and four normal nasopharyngeal tissues.7 The results suggest that the carcinogenesis of NPC could be ascribed to a loss of miR-34b, which raises questions regarding the cause of the downregulation of miR-34b. An increasing number of studies have proposed that the expression of miR-34b is inversely correlated with CpG island methylation.28 In addition to epigenetic regulation, miR-34b is also a target gene of p53.29–31 Because p53 mutations are rare in NPC, whether miR-34b is regulated by p53 warrants further investigation.
USP22 belongs to the ubiquitin hydrolase family and has recently generated a large amount of attention due to its association with cancer. A microarray analysis revealed that a high level of USP22 is commonly seen in many types of cancers.10 In oral squamous cell carcinoma and cervical cancer cells, USP22 was identified as a novel molecular biomarker that is predictive of cancer progression and patient prognosis.11,12 However, the expression of USP22 in NPC has remained unclear. Here, we compared the expression of USP22 in NPC cell lines and immortalized nasopharyngeal epithelial cells. The data showed that USP22 was overexpressed in NPC cell lines when compared with NP69. Accentuated USP22 expression in many types of cancer cells usually indicates a poor clinical outcome. The influence of USP22 expression on the clinical prognosis of NPC requires further analysis. Recently, USP22 was identified as a subunit of the Spt–Ada–Gcn5 acetyltransferase coactivator complex, which regulates the expression of genes that are related to oncogenicity and proliferation.32 We assessed the function of USP22 in NPC proliferation. The overexpression or knockdown of USP22 promoted the proliferation of NPC. Notably, the overexpression of USP22 rescued the inhibitory effect of miR-34b on NPC, demonstrating that miR-34b suppresses the proliferation of NPC by targeting USP22.
Conclusion
In summary, our recent study showed that miR-34b was expressed at a low level in NPC and acted as positive regulator of NPC proliferation. By contrast, USP22 was overexpressed in NPC cells and promoted the proliferation of NPC. Furthermore, we demonstrated that miR-34b inhibited NPC proliferation by downregulating the expression of USP22. The results of this study provide a theoretical basis for miR-34b- or USP22-targeted diagnoses and therapies for NPC.
Acknowledgments
This work was supported by grants from the Guangdong Natural Science Foundation (No S2012040007005), the National Natural Science Foundation of China (No 81274145), and the Ministry of Education, Science Technology Development Center (No 20124425120012).
Disclosure
The authors report no conflicts of interest in this work.
References
Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma – review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30(7):946–963. | ||
Wei WI, Sham JS. Nasopharyngeal carcinoma. Lancet. 2005;365(9476):2041–2054. | ||
Yoshizaki T, Ito M, Murono S, Wakisaka N, Kondo S, Endo K. Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx. 2012;39(2):137–144. | ||
Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. | ||
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. | ||
He ML, Luo MX, Lin MC, Kung HF. MicroRNAs: potential diagnostic markers and therapeutic targets for EBV-associated nasopharyngeal carcinoma. Biochim Biophys Acta. 2012;1825(1):1–10. | ||
Li T, Chen JX, Fu XP, et al. microRNA expression profiling of nasopharyngeal carcinoma. Oncol Rep. 2011;25(5):1353–1363. | ||
Chen HC, Chen GH, Chen YH, et al. MicroRNA deregulation and pathway alterations in nasopharyngeal carcinoma. Br J Cancer. 2009;100(6):1002–1011. | ||
Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns. 2006;6(3):277–284. | ||
Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5(11):1208–1216. | ||
Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumour Biol. 2014;35(2):929–934. | ||
Piao S, Liu Y, Hu J, et al. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS One. 2012;7(8):e42540. | ||
Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ, Jiang GS. Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol Cell Biochem. 2011;346(1–2):11–21. | ||
Ling SB, Sun DG, Tang B, et al. Knock-down of USP22 by small interfering RNA interference inhibits HepG2 cell proliferation and induces cell cycle arrest. Cell Mol Biol (Noisy-le-grand). 2012;58(suppl):L1803–L1808. | ||
Zhang XY, Varthi M, Sykes SM, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29(1):102–111. | ||
Liu X, Lv XB, Wang XP, et al. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11(13):2495–2506. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Spano JP, Busson P, Atlan D, et al. Nasopharyngeal carcinomas: an update. Eur J Cancer. 2003;39(15):2121–2135. | ||
Marquitz AR, Raab-Traub N. The role of miRNAs and EBV BARTs in NPC. Semin Cancer Biol. 2012;22(2):166–172. | ||
Yang S, Li Y. MicroRNAs: novel factors in clinical diagnosis and prognosis for nasopharyngeal carcinoma. Acta Pharmacol Sin. 2012;33(8):981–982. | ||
Liu X, Luo HN, Tian WD, et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol Ther. 2013;14(12):1133–1142. | ||
Liu N, Tang LL, Sun Y, et al. MiR-29c suppresses invasion and metastasis by targeting TIAM1 in nasopharyngeal carcinoma. Cancer Lett. 2013;329(2):181–188. | ||
Cai K, Wan Y, Sun G, Shi L, Bao X, Wang Z. Let-7a inhibits proliferation and induces apoptosis by targeting EZH2 in nasopharyngeal carcinoma cells. Oncol Rep. 2012;28(6):2101–2106. | ||
Yi C, Wang Q, Wang L, et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31(41):4421–4433. | ||
Zhang LY, Ho-Fun LV, Wong AM, et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34(2):454–463. | ||
Svoboda M, Sana J, Redova M, et al. MiR-34b is associated with clinical outcome in triple-negative breast cancer patients. Diagn Pathol. 2012;7:31. | ||
Harata K, Ishiguro H, Kuwabara Y, et al. MicroRNA-34b has an oncogenic role in esophageal squamous cell carcinoma. Oncol Lett. 2010;1(4):685–689. | ||
Wong KY, Yu L, Chim CS. DNA methylation of tumor suppressor miRNA genes: a lesson from the miR-34 family. Epigenomics. 2011;3(1):83–92. | ||
Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014;6(3):214–230. | ||
Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67(18):8433–8438. | ||
Li L, Wu J, Sima X, et al. Interactions of miR-34b/c and TP-53 polymorphisms on the risk of nasopharyngeal carcinoma. Tumour Biol. 2013;34(3):1919–1923. | ||
Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7(11):1522–1524. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.