Back to Journals » OncoTargets and Therapy » Volume 11
MiR-216b suppresses colorectal cancer proliferation, migration, and invasion by targeting SRPK1
Received 7 January 2018
Accepted for publication 6 February 2018
Published 23 March 2018 Volume 2018:11 Pages 1671—1681
DOI https://doi.org/10.2147/OTT.S161835
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ingrid Espinoza
Yanfen Yao,1 Qiaorong Li,1 Hong Wang2
1Department of Intensive Care Unit, Shandong Provincial Third Hospital, Jinan, People’s Republic of China; 2Department of General Surgery, Shandong Provincial Third Hospital, Jinan, People’s Republic of China
Background: MiR-216b has been reported to be involved in the development of some cancers, however, the role of miR-216b in colorectal cancer (CRC) remains unclear.
Purpose: This study aimed to investigate the mechanism underlying miR-216b-induced CRC development.
Methods: We detected the expression of miR-216b in 80 cases of CRC tissues and cell lines, and further analyzed the association between miR-216b and clinical pathological indicators as well as prognosis. In vitro, the miR-216b overexpression cell model was established for further functional assay.
Results: We demonstrated that miR-216b in CRC tissues and cell lines was markedly decreased compared with corresponding adjacent normal tissues and colonic mucosal epithelial cell line, and was obviously associated with the TNM stage, lymph node metastases, and poor overall survival as well as recurrence-free survival. Furthermore, we found that miR-216b inhibited cell proliferation, cell cycle, migration, and invasion by targeting 3'-UTR of SRPK1. Besides, SRPK1 over-expression reversed miR-216b-inhibited cell proliferation, migration and invasion, while SRPK1 inhibition aggravated these effects.
Conclusions: We identified that miR-216b suppresses colorectal cancer proliferation, migration and invasion by targeting SRPK1, which shed light on how miR-216b functions in CRC pathogenesis.
Keywords: MiR-216b, SRPK1, colorectal cancer, proliferation, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most common malignant cancers, with approximately one million patients being diagnosed every year.1 Of these CRC patients, nearly 20% develop distant metastasis, which often results in a poor prognosis.2,3 The current therapy for CRC is surgery combined with radiochemotherapy. However, despite therapy, a subset of patients are accompanied by local recurrences and metastases from tumor sites.4,5 Thus, it is essential to hunt for a new target to repress CRC development.
MicroRNAs (miRNAs) are a kind of small noncoding RNAs, which regulate mRNA expression at the posttranscriptional level by cleaving mRNA and/or repressing translational processes.6–8 miRNAs are involved in both physiological and pathological processes, such as cell proliferation, cell cycle arrest, development, metabolism, and oncogenesis.9,10 Increasing evidence suggests that miR-216b serves as either a tumor suppressor or an oncogene by targeting relevant mRNAs involved in tumor development and progression in some cancers, and hence is used for potential intervention.11–13 In view of these functions, the expression of miR-216b can be downregulated in malignant cells by chemical modification of oligonucleotides, which alters cancer phenotypes.14–16 However, few studies on miR-216b expression and CRC development are found in recent decades.
In a previous study, we demonstrated that SRPK1 plays an oncogenic role in gastric cancer development.17 In addition, we found that SRPK1 is a potential target of miR-216b by bioinformatic analysis. In this study, we explored whether miR-216b is involved in CRC progression by targeting SRPK1 and the specific underlying mechanisms. We detected the expression of miR-216b in 80 CRC tissues and cell lines, and further analyzed the association between miR-216b and clinicopathological indicators as well as prognosis in 60 cases. We also established the miR-216b overexpression cell model in vitro for further functional assay.
Materials and methods
Specimen collection
A total of 80 CRC specimens, including adjacent nontumor tissues, were obtained from patients in the Shandong Provincial Third Hospital and Shandong Cancer Hospital between March 2011 and February 2012. Adjacent nontumor tissues located at least 5 cm from tumors were used as controls. The Institutional Ethics Committee of Shandong Provincial Third Hospital approved the study protocol and use of clinical specimen. Written, voluntary, informed consent was taken from all the patients. None of the patients received chemotherapy or radiotherapy before the surgery. The resected tissues were immediately snap frozen in liquid nitrogen and stored until RNA extraction. Histological grade was determined blindly by two pathologists. Of the total 80 patients, 60 were followed up for 5 years and clinical assessment was performed in each patient at the end of study.
Cell lines
Colonic mucosal epithelial cell line (FHC) and CRC cell lines (HCT8 and HT29) were obtained from the Chinese Academy of Medical Sciences (Beijing, People’s Republic of China). Cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; HyClone) in a humidified incubator under 5% CO2 at 37°C.
Transfection
Cell transfection was conducted in cell lines using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer’s instructions. Briefly, cells were cultured to 70% confluence, and resuspended in serum-free DMEM. Scramble miR (miR-NC), miR-216b mimic for miR-216b upregulation, and pc-DNA3.1-SRPK1 plasmid for SRPK1 overexpression were diluted in OPTI-MEM (Thermo Fisher Scientific), and the mixture was then added with diluted Lipofectamine 2000. All mimics and plasmids were purchased from Yearthbio (Changsha, People’s Republic of China). Following incubation for 20 min at room temperature, the mixture was added into the cell suspension. Following incubation at 37°C under 5% CO2 for 6 h, the transfection mixture was replaced with DMEM with 10% FBS. MiR-NC was the corresponding control for miR-216b mimics. For SRPK1 silencing, SRPK1 siRNA lentiviral vectors were constructed by inserting SRPK1 sequences into the GV248 lentiviral vector (GeneChem Company, Shanghai, People’s Republic of China). Then, the viral supernatants were collected and filtered 48 h after the transfection. HCT8 and HT29 cells were infected with the viral supernatant, and successfully infected cells were selected using puromycin (0.5 μg/mL) (#P8833; Sigma-Aldrich Co., St Louis, MO, USA). Finally, the expression of SRPK1 was confirmed by Western blot.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA, including miRNA, was isolated from tissues or cell lines using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. For miRNA expression analysis, reverse transcription was performed using the TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) with miR-216b-specific primers (Applied Biosystems). Mature miR-216b levels were quantified with TaqMan miRNA assays (Applied Biosystems). For SRPK1 mRNA detection, reverse transcription was performed using the PrimeScript RT reagent kit (Takara, Tokyo, Japan). Quantitative PCR was performed using SYBR Premix Ex Taq (Takara) on the ABI 7500 real-time PCR System (Applied Biosystems). U6 snRNA or GAPDH was used as internal control. The relative expression levels were calculated by the equation 2−ΔΔCT.
Western blot analysis
The cells were suspended in radioimmunoprecipitation assay protein lysis buffer (pH 7.4). Protein concentration was quantified using BCA protein assay kit (Thermo Fisher Scientific). Total protein (50 μg) was resolved with 6% SDS-PAGE, and transferred to a polyvinylidene difluoride membrane. Membranes were blocked in 5% nonfat milk diluted in TBST for 1 h at room temperature. The blots were probed with primary antibodies against SRPK1 (dilution, 1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). After washing, membranes were probed with appropriate secondary antibodies, horseradish peroxidase-conjugated goat anti-mouse IgG (dilution 1:5,000; Boster Biological Technology, Pleasanton, CA, USA). Secondary antibodies were incubated at room temperature for 1 h. All membranes were stripped by incubating in Restore PLUS Western blot stripping buffer (Thermo Fisher Scientific) for 15 min at room temperature and reprobed with anti-GAPDH antibody for control. The blots were visualized using enhanced chemiluminescence (GE Healthcare, Little Chalfont, UK).
Cell counting kit-8 (CCK-8) assay
Cells were seeded into 96-well plates at 5×103 cells per well. After incubation overnight, cell transfection was performed and then the cells were incubated at 37°C in humidified air with 5% CO2. Cell proliferation was examined at 0, 24, 48, and 72 h after transfection. Briefly, 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan) was added into each well and incubated at 37°C for another 2 h. Finally, the optical density was detected at a wavelength of 450 nm using the ELISA plate reader (Model 550; Bio-Rad Laboratories, Hercules, CA, USA). At least three independent experiments were performed.
FACS (fluorescence-activated cell sorter) analysis
After transfection for 24 h, cells were first fixed in ice-cold 70% ethanol and then stained with PBS containing propidium iodide (50 μg/mL)/Annexin V and RNase A (100 μg/mL) for DNA analysis using flow cytometry on a BD FACSCalibur system (FACScan; BD Biosciences, San Diego, CA, USA). Percentage of the cells in the different phases of cellular cycle was measured using the CellQuest software version 3.3 (Becton Dickinson Immunocytometry Systems, San Jose, CA, USA).
Transwell assay
Transwell assays were performed in 24-well transwell plates of 8 μm pore size according to the manufacturer’s instructions (Corning, Corning, NY, USA). The bottom of transwell chamber was coated with BD Matrigel Basement Membrane Matrix. The upper chamber was filled with 1×105 cells in RPMI 1640 containing 5% FBS. The lower chamber was filled with RPMI 1640 containing 25% FBS as a chemoattractant. After the chambers were incubated for 24 or 48 h at 37°C, non-invading cells on the upper side of the chamber were removed from the surface of the membrane by scrubbing, and invading cells on the lower surface of the membrane were fixed with methanol, mounted, and dried. The number of cells through the matrigel was counted by a technician blinded to the experimental settings in four randomly selected microscopic fields of each filter. The test was conducted in three biological replicates. The results were normalized to a blank (0 h).
Luciferase reporter assay
To predict the potential targets of miR-216b, bioinformatic analysis was performed with TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/getExprForm.do).
The pMIR-SRPK1-3′-UTR-wild-type and pMIR-SRPK1-3′-UTR-mutant containing the putative binding site of miR-216b were synthesized and confirmed by GenePharma, Co., Ltd. (Shanghai, People’s Republic of China). Cells were seeded into 24-well plates and cultured until the cell density reached 80%–90% confluence. Subsequently, cells were transfected with either the pMIR-SRPK1-3′-UTR-WT or the pMIR-SRPK1-3′-UTR-MUT reporter vector, together with miR-216b mimics or miR-NC, using Lipofectamine 2000. After incubation for 48 h, the activities of firefly and Renilla luciferases were determined in transfected cells using the dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s recommendations. Renilla-luciferase activity was assayed for normalization.
Statistical analysis
Statistical tests were carried out using GraphPad Prism 5. The differences between groups were analyzed using Student’s t-test when only two groups were compared or one-way analysis of variance when more than two groups were compared. Recurrence-free survival (RFS) was defined as the time from diagnosis of CRC to first locoregional or distant recurrence. Overall survival (OS) was the time from CRC diagnosis to death. RFS and OS curves were calculated using the Kaplan–Meier method and compared by log-rank testing. p<0.05 was considered statistically significant.
Results
MiR-216b expression is downregulated in CRC tissues and cells, and correlates with SRPK1 expression
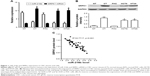
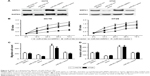
Firstly, we detected the expression of miR-216b using qRT-PCR in CRC tissues and paired noncancerous tissues as well as cell lines. We found that miR-216b expression levels were about 37% lower in CRC cancer samples than that in paired noncancerous tissues (p<0.05; Figure 1A). Similarly, the expression level of miR-216b was also markedly decreased by about 64% in HCT8, and by about 68% in HT29 cell lines than that in FHC cells (p<0.05; Figure 1A).
Next, we carried out RT-qPCR and Western blot to detect the expression of SRPK1 in CRC tissues and normal tissues. We found that the expression of SRPK1 mRNA and protein was ~4.7-fold and 6.1-fold higher in CRC tissues compared with normal tissues (p<0.05; Figure 1A and B). Furthermore, the expression of SRPK1 mRNA and protein was also significantly upregulated in HCT8 (4.6-fold and 5.3-fold) and HT29 (4.8-fold and 5.3-fold) cells, compared with FHC cells (p<0.05; Figure 1A and B). Importantly, we demonstrated an inverse correlation between miR-216b and SRPK1 protein expression in CRC tissues (R2=0.7717, p<0.001; Figure 1C).
Association of miR-216b and SRPK1 expression with clinical data
To evaluate the expression and significance of miR-216b and SRPK1 in CRC tissues, the expression of miR-216b and SRPK1 was classified as low and high based on the median of miR-216b and SRPK1 expression levels. The clinicopathological significance of miR-216b and SRPK1 expression in CRC patients is shown in Table 1. Notably, low miR-216b and SRPK1 level correlated with lymph node metastasis and TNM stage (all p<0.05).
  | Table 1 Relationship of miR-216b and SRPK1 with clinicopathological indicators |
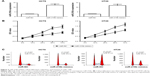
To further assess the prognostic significance of miR-216b and SRPK1 expression in CRC, Kaplan–Meier survival curve was constructed to analyze the survival outcome of CRC patients. OS analysis revealed that CRC patients with low miR-216b or high SRPK1 expression had a poorer OS compared with those with high miR-216b (log-rank test, p=0.0386; Figure 2A) or low SRPK1 expression (log-rank test, p=0.0013; Figure 2B). RFS analysis revealed that CRC patients with low miR-216b or high SRPK1 expression had a poorer RFS compared with those with high miR-216b (log-rank test, p=0.0295; Figure 2C) or low SRPK1 expression (log-rank test, p=0.0219; Figure 2D). Additionally, multivariate Cox regression analysis indicated that miR-216b and SRPK1 (mRNA and protein) expression was an independent risk factor for poor prognosis of CRC patients (p=0.024, p=0.009, and p=0.012, respectively; Table 2). Thus, miR-216b and SRPK1 expression potentially acts as an independent predictor for prognosis of CRC patients.
  | Table 2 Univariate and multivariate Cox regression analysis of overall survival duration |
Ectopic expression of miR-216b inhibited the proliferation of CRC cells
In order to investigate the role of miR-216b in cell proliferation, miR-216b mimics were transfected into HCT8 and HT29 cell lines. Quantitative real-time PCR confirmed the increased expression of miR-216b after transfection (Figure 3A). CCK-8 assay showed that ectopic expression of miR-216b obviously inhibited the proliferation of HCT8 and HT29 cells compared with their control at 48 h (both p<0.05; Figure 3B). As reported, the inhibition of proliferation may be partially attributed to cell cycle arrest. Subsequently, we investigated the effect of miR-216b on cell cycle regulation. At 48 h post-transfection, cell cycle analysis revealed that ectopic expression of miR-216b significantly induced cell cycle arrest in the G0/G1 phase in HCT8 and HT29 cells compared with the control group (both p<0.05; Figure 3C).
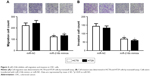
MiR-216b affects migration and invasion of CRC cells
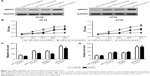
We further investigated the effect of miR-216b on cell migration and invasion using the transwell assay after transfection with miR-216b mimics. We found that miR-216b overexpression markedly decreased the migration and invasive ability of HCT8 and HT29 cells compared with the control group (both p<0.05; Figure 4A). Transwell assay further showed the upregulation of miR-216b significantly reduced the invasive ability of HCT8 and HT29 cells compared with the control group (both p<0.05; Figure 4B).
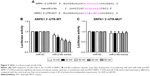
MiR-216b regulates SRPK1 expression by directly targeting its 3′-UTR
To investigate the target genes of miR-216b, the potential targets of miR-216b were predicted using TargetScanHuman 6.2. Our analysis showed that SRPK1 was a potential target of miR-216b (Figure 5A). To determine whether miR-216b directly targets the SRPK1 3′-UTR, the luciferase reporter assay was conducted. HCT8 cells were co-transfected with luciferase reporter plasmids and miR-216b mimics or miR-NCs. We demonstrated that miR-216b overexpression obviously inhibited the luciferase activity of HCT8 cells containing the wild-type SRPK1 3′-UTR (Figure 5B), while miR-216b mimics did not affect the luciferase activity of the reporter gene containing the mutated SRPK1 3′-UTR compared with their respective controls (Figure 5C). Furthermore, we used Western blot analysis to identify the impact of miR-216b on SRPK1 expression. We found that overexpression of miR-216b obviously reduced the expression of SRPK1 protein in HCT8 and HT29 cells compared with miR-NC group (both p<0.05; Figure 6A), indicating that miR-216b repressed SRPK1 expression at the posttranscriptional level.
SRPK1 mediates the biological functions of miR-216b in CRC cells
To identify whether SRPK1 could regulate the biological processes of miR-216b in CRC cells, HCT8 and HT29 cells were transfected with miR-216b mimics, miR-NC, SRPK1 plasmids, or vectors. Transfection of SRPK1 plasmids significantly increased the protein level of SRPK1 in HCT8 and HT29 cells. We further investigated the role of SRPK1 overexpression in miR-216b-mediated CRC proliferation, migration, and invasion. It was found that SRPK1 overexpression obviously reversed miR-216b-inhibited HCT8 and HT29 proliferation (p<0.05; Figure 6B). Furthermore, the transwell assay revealed that SRPK1 overexpression also promoted miR-216b-inhibited cell migration and invasion compared with HCT8 and HT29 cells co-transfected with miR-216b mimics and vectors (p<0.05; Figure 6C and D). Afterward, we carried out SRPK1 silencing assay, and transfected SRPK1 siRNA and control siRNA into HCT8 and HT29 cells with miR-216b mimics, and further investigated the role of SRPK1 in miR-216b-mediated proliferation, migration, and invasion (Figure 7A). We found that si-SRPK1 obviously aggravated miR-216b-inhibited HCT8 and HT29 proliferation, migration, and invasion (p<0.05; Figure 7B–D). These results suggest that SRPK1 mediates the biological functions of miR-216b in CRC cells.
Discussion
Emerging reports showed that miRNAs play an important role in biological processes of different cancers, including cell proliferation, apoptosis, invasion, and migration.12–15 Recent findings revealed several miRNAs, such as miR-105, miR-193b, miR-1207-5p, and so on, were involved in the tumorigenesis and progression of CRC.18–20 miR-216b serves as either a tumor suppressor or an oncogene by targeting relevant mRNAs in tumor development and progression in some cancers, and hence is used for potential intervention.11–13 In view of these functions, the expression of miR-216b can be downregulated in malignant cells by chemical modification of oligonucleotides, which alters cancer phenotypes.14–16 However, the direct role of miR-216b in CRC remains elusive.
In this study, we observed that miR-216b was significantly decreased in CRC tissues compared with nontumor tissues. Additionally, miR-216b expression was also downregulated in CRC cell lines. Consistent with our data, Chen et al demonstrated that the expression of miR-216b in CRC tissues and cell lines was markedly decreased compared with corresponding adjacent normal tissues and FHC, and miR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling pathway. These results suggest that miR-216b may also exert an inhibitory effect on the growth and metastasis of CRC.16
miRNAs exert their biological function by regulating specific target genes. To elucidate the mechanisms underlying miR-216b-inhibited CRC progression, we selected potential targets of miR-216b in CRC cells. Of candidate target genes, we found SRPK1 to be an oncogene, which was previously reported to be involved in the progression of CRC. It has been reported that SRPK1 was highly expressed in many kinds of cancers, including prostate cancer, breast cancer, lung cancer, and glioma.17,21 A couple of reports demonstrated that inhibition of SRPK1 exerts antitumor effects on different tumors; therefore, SRPK1 has been recommended as a novel candidate for cancer patient therapies.22,23 In this study, SRPK1 expression was found to be significantly increased in CRC tissues compared with nontumor tissues, and its expression negatively correlated with miR-216b expression in CRC tissues. Besides, the luciferase reporter assay indicated that miR-216b directly bound to the 3′-UTR of SRPK1 mRNA, and repressed SRPK1 expression at the posttranscriptional level. Most importantly, upregulation of SRPK1 markedly reversed the inhibitory effects of miR-216b on the proliferation and invasion of CRC cells, while inhibition of SRPK1 aggravated the inhibitory effects of miR-216b. These results indicated that SRPK1 indeed acts as a downstream effector in miR-216b-repressed CRC proliferation and invasion.
In conclusion, our study shows that miR-216b is an important metastasis-related miRNA and significantly downregulated in CRC tissues. Overexpression of miR-216b markedly inhibits the proliferation, migration, and invasion of CRC cells by targeting SRPK1 expression.
Acknowledgment
The authors are thankful to other members in their laboratory for their suggestions.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. | ||
Scurr M, Pembroke T, Bloom A, et al. Effect of modified vaccinia ankara-5T4 and low-dose cyclophosphamide on antitumor immunity in metastatic colorectal cancer: a randomized clinical trial. JAMA Oncol. 2017;3(10):e172579. | ||
Singal AG, Gupta S, Skinner CS, et al. Effect of colonoscopy outreach vs fecal immunochemical test outreach on colorectal cancer screening completion: a randomized clinical trial. JAMA. 2017;318(9):806–815. | ||
Rat C, Pogu C, Le Donné D, et al. Effect of physician notification regarding nonadherence to colorectal cancer screening on patient participation in fecal immunochemical test cancer screening: a randomized clinical trial. JAMA. 2017;318(9):816–824. | ||
Tanaka H, Hazama S, Iida M, et al. miR-125b-1 and miR-378a are predictive biomarkers for the efficacy of vaccine treatment against colorectal cancer. Cancer Sci. 2017;108(11):2229–2238. | ||
Slaby O, Laga R, Sedlacek O. Therapeutic targeting of non-coding RNAs in cancer. Biochem J. 2017;474(24):4219–4251. | ||
Xu-Welliver M, Carbone DP. Blood-based biomarkers in lung cancer: prognosis and treatment decisions. Transl Lung Cancer Res. 2017;6(6):708–712. | ||
Pereira CM, Sehnem D, da Fonseca EO, et al. miRNAs: important targets for oral cancer pain research. Biomed Res Int. 2017;2017:4043516. | ||
Moretti F, D’Antona P, Finardi E, et al. Systematic review and critique of circulating miRNAs as biomarkers of stage I-II non-small cell lung cancer. Oncotarget. 2017;8(55):94980–94996. | ||
Karmakar S, Kaushik G, Nimmakayala R, et al. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. Epub 2017 Dec 2. | ||
Liu FY, Zhou SJ, Deng YL, et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2015;6:e1670. | ||
Endo K, Weng H, Kito N, et al. MiR-216a and miR-216b as markers for acute phased pancreatic injury. Biomed Res. 2013;34(4):179–188. | ||
Deng M, Tang H, Zhou Y, et al. miR-216b suppresses tumor growth and invasion by targeting KRAS in nasopharyngeal carcinoma. J Cell Sci. 2011;124(Pt 17):2997–3005. | ||
Liu Y, Niu Z, Lin X, et al. MiR-216b increases cisplatin sensitivity in ovarian cancer cells by targeting PARP1. Cancer Gene Ther. 2017;24(5):208–214. | ||
Sun M, Wang X, Tu C, et al. microRNA-216b inhibits cell proliferation and migration in human melanoma by targeting FOXM1 in vitro and in vivo. Cell Biol Int. 2017;41(12):1272–1282. | ||
Chen X, Liu X, He B, et al. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7(10):2051–2069. | ||
Wang H, Wang C, Tian W, et al. The crucial role of SRPK1 in IGF-1-induced EMT of human gastric cancer. Oncotarget. 2017;8(42):72157–72166. | ||
Shen Z, Zhou R, Liu C, et al. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213. | ||
Liu KL, Luo JL, Wu J, Wang YD, Fan HJ. A preliminary study of the value of plasma microRNA-193b and soluble urokinase-type plasminogen activator receptor in identifying patients with early-stage colorectal cancer and. Clin Lab. 2017;63(11):1949–1953. | ||
Wang X, Wu X. The role of microRNA-1207-5p in colorectal cancer. Clin Lab. 2017;63(11):1875–1882. | ||
Batson J, Toop HD, Redondo C, et al. Development of potent, selective SRPK1 inhibitors as potential topical therapeutics for neovascular eye disease. ACS Chem Biol. 2017;12(3):825–832. | ||
Claessens A, Affara M, Assefa SA, Kwiatkowski DP, Conway DJ. Culture adaptation of malaria parasites selects for convergent loss-of-function mutants. Sci Rep. 2017;7:41303. | ||
Bullock N, Oltean S. The many faces of SRPK1. J Pathol. 2017;241(4):437–440. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.