Back to Journals » OncoTargets and Therapy » Volume 13
miR-195 Serves as a Tumor Suppressor in the Progression of Liposarcoma by Targeting OSBP
Authors Cao Y, Li L, Han L, Zheng J, Lv C
Received 16 December 2019
Accepted for publication 23 May 2020
Published 6 July 2020 Volume 2020:13 Pages 6465—6474
DOI https://doi.org/10.2147/OTT.S242608
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Ye Cao,1,* Lei Li,1,* Lu Han,2 Jiajia Zheng,3 Chentao Lv1
1Department of General Surgery, Shanghai Public Health Clinical Center, Fudan University, Shanghai, People’s Republic of China; 2Department of General Surgery, Shanghai Jiangong Hospital, Shanghai, People’s Republic of China; 3Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiajia Zheng
Department of General Surgery, Zhongshan Hospital, Fudan University, No. 180, Fenglin Road, Xuhui District, Shanghai 200032, People’s Republic of China
Tel +86-21-64041990
Email [email protected]
Chentao Lv
Department of General Surgery, Shanghai Public Health Clinical Center, Fudan University, No. 2901, Caolang Highway, Jinshan District, Shanghai 200083, People’s Republic of China
Tel +86-21-37990333
Email [email protected]
Background: Liposarcoma was considered as a soft tissue kind of sarcoma with one-fifth in the sarcoma cases of adults. The aim of this study was to explore the role and the potential mechanisms of miR-195 in liposarcoma.
Methods: Quantitative real-time PCR (qRT-PCR) was conducted to measure the expression of microRNA-195 (miR-195) and oxysterol-binding protein (OSBP) in liposarcoma. Cell proliferation was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT). Cell migration was measured by wound healing and transwell assays. Cell cycle phases and apoptosis were examined using flow cytometry analysis. Caspase-3 activity was detected by commercial kit. Binding sites between miR-195 and OSBP were predicted through bioinformatics analysis and confirmed by dual-luciferase reporter assay and RNA immunoprecipitation (RIP). Western blot was used to analyze OSBP level. Xenograft tumor assays were performed to observe the effect of miR-195 overexpression on tumor growth in vivo.
Results: miR-195 expression was decreased, whereas OSBP was increased in liposarcoma tissues and cells. Besides, miR-195 upregulation suppressed the proliferative and migrative abilities and induced inhibition on cell growth and promotion on apoptosis in SW872 and 93T449 cells. Mechanically, miR-195 functioned as a suppressor by regulating OSBP expression. Furthermore, OSBP overexpression inverted the effects of miR-195 on cell growth, migration and apoptosis in SW872 and 93T449 cells. miR-195 overexpression also suppressed tumor growth in vivo.
Conclusion: miR-195 suppressed cell growth, migration and elevated cell apoptosis via OSBP in liposarcoma.
Keywords: liposarcoma, miR-195, OSBP, proliferation, migration, cell cycle, apoptosis
Introduction
As we know, liposarcoma was regarded as a soft tissue type of sarcoma.1 Other than primary local disease could be effectively resected by surgery, while the operating method was not easy to remedy in the cases of metastatic disease, but chemotherapy or radiotherapy was replenishable. The occurrence rate of liposarcoma was high, and the molecular mechanisms were few understood by researchers. Liposarcoma was mainly classified into triple subtypes relying on their histopathological profiles,2,3 namely well-differentiated/de-differentiated liposarcoma, myxoid/round cell liposarcoma and pleomorphic liposarcoma. The first type of liposarcoma was resistant to conventional chemotherapy or radiotherapy. Thus, it is meaningful to explore the novel targets for the treatment of liposarcoma.
Following the exploration for oncogenes and cancer-suppressors, in recent years, small RNAs including microRNAs (miRNAs) have become novel molecules that inhibited cancer progression. miRNAs, with 20–24 nucleotides, could bind to the 3ʹ untranslated region (3ʹUTR) of mRNAs to restrain the progression of translation and mRNA stability.4–6 Candidate miRNAs have functioned to mediate hundreds of transcripts, either directly or indirectly,7 thereby achieving pleiotropic control of cellular processes. Dysregulated expression of miRNAs existed in multiple cancers, such as lung cancer, breast cancer, prostate cancer, and gastrointestinal cancers.8,9
Disordered miRNAs seemed to be regarded as tumor suppressors. It was widely reported that restored miRNAs expression can usually be regarded as tumor therapeutic agents. Other miRNAs seemed to be oncogenes,10 so anti-miRNAs11 could be considered as a therapeutic for cancers that overexpress the miRNA. In Casadei et al study, he predicted that miR-25-3p and miR-92a-3p may be useful as potential biomarkers of liposarcoma, due to the evidence of secretion through extracellular vesicles.12 Yu et al uncovered low-expression of miR-133a in human dedifferentiated liposarcoma tissues and cells. Enforced miR-133a expression decreased proliferative and glycolytic abilities, impacted cell cycle progression kinetics, and increased oxidative phosphorylation.13 In the past few years, miR-195 served as a tumor suppressor in diversiform cancers, such as hepatocellular carcinoma,14 colorectal cancer,15 lung adenocarcinoma,16 pancreatic cancer.17 Yu et al showed that miR-195 overexpression inhibited the proliferation, migration, invasion and lipogenesis in pancreatic cancer cells.17 Chae et al investigated that miR-195 inhibited SMAD Specific E3 Ubiquitin Protein Ligase 2 (SMURF2) mRNA expression by binding to its 3ʹ-UTR. In lung cancer cells, miR-195 mediated SMURF2-dependent TβRI ubiquitination and activated the transforming growth factor-β (TGF-β) pathway. Also, miR-195 enhanced level reduced viability, proliferation, and invasion via TGF-β signaling activation. In vivo studies of xenograft model revealed the repressed-effect of miR-195 on tumor growth.18 More importantly, a previous study identified that miR-195 was downregulated in liposarcoma tissues.19 Thus, it was needed to explore the character and potential mechanism of miR-195 in liposarcoma.
The eukaryotic gene family consisting of oxysterol-binding protein (OSBP) and OSBP-related protein (ORP) were involved in the processes of transporting and metabolism of sterols and phospholipids.20 OSBP, as the founder member of the ORP family, was identified as a soluble receptor for oxysterols (oxidized cholesterol derivatives).21,22 OSBP played an essential part in cell signaling pathways,23 as well as shared a characteristic feature (conserved C-terminal OSBP-related ligand-binding (ORD) domain)24 with ORPs. Emerging evidence suggested that OSBP appeared to participate in cancer evolution. For example, OSBP knockdown apparently blocked cell growth and AKT phosphorylation in SW872-S cells.25 However, the function of OSBP in liposarcoma was currently unclear.
In this research, we probed the profile of miR-195 and OSBP in liposarcoma, bio-functional effects and underlying mechanisms of which in liposarcoma.
Materials and Methods
Clinical Samples
Liposarcoma or normal fat tissues were obtained from patients (N=28) or randomly selected healthy volunteers (N=8) at Shanghai Public Health Clinical Center, Fudan University. The Ethics Committee of Shanghai Public Health Clinical Center, Fudan University approved the protocol of this study, and written informed consent was signed by all participants.
Cell Culture and Transfection of Oligonucleotide and Plasmid
Human SW872 and 93T449 cells (American Type Culture Collection, ATCC, Manassas, VA, USA) were grown in Dulbecco’s Modified Eagle Medium (Gibco, Carlsbad, CA, USA) containing 10% fetal bovine serum and 1% antibiotics (TaKaRa, Dalian, China). All cells were maintained in a 37°C, 5% CO2 incubator. miR-195 (GenePharma, Shanghai, China), and its corresponding scrambled oligonucleotide sequence (miR-con, GenePharma), anti-miR-195 (GenePharma) and its corresponding scrambled oligonucleotide sequence (anti-miR-con, GenePharma) were transfected into cells using Dharma FECT 4 transfection reagent (Dharmacon, Brebières, France), referring to the protocols of manufacturers. The full sequences of OSBP were inserted into the pcDNA vector (Promega, Southampton, UK) to construct OSBP overexpression plasmid (OSBP), and nontarget vector (pcDNA) was used as a negative control.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
A commercial Total RNA Purification Kit (Norgen Biotek, Thorold, Canada) was used to extract total RNA from tissues and cells. Total RNA (1 µg) was used for cDNA synthesis with Omniscript Reverse Transcriptase (Qiagen, Hombrechtikon, Switzerland) for OSBP or miScript Reverse Transcription Kit (Qiagen) for miR-195. Synthetic cDNA was then subjected to qRT-PCR using FastStart SYBR Green Master (Roche, Laval, Canada) on the Roche Light Cycler 480 System. The relative fold change was calculated by the 2−ΔΔCt method with β-actin or U6 for normalization. The following primers (5ʹ to 3ʹ) were as followed: miR-195-Forward: GGGGAGCCAAAAGGGTCATCATCT and miR-195-Reverse: GAGGGGCCATCCACAGTCTTCT, OSBP-Forward: AGATCCCCATGCCGGTAAAC and OSBP-Reverse: CGTCCCCAGAAGAGCAAAGT, β-actin-Forward: GGACAAGGAGGGACCTCTTC and β-actin-Reverse: GAACATGAGA CACACCTAGC, U6-Forward: CTCGCTTCGGCAGCACA and U6-Reverse: AACGCTTCACGAATTTGCGT.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT)
SW872 and 93T449 cells were plated in 96-well cell culture plates. A 20 μL 5 mg/mL MTT reagent (Sigma, St Louis, MO, USA) was added into the supernatant of cells. A549 cells were incubated with MTT in the incubator at 37°C with 5% CO2 for 4 h. The culture medium was discarded, and DMSO (Sigma) was used to dissolve formazan crystals. Absorbance of each well at 490 nm was examined using a microplate reader.
Wound Healing
Wound healing assays were performed to determine cell migration. A total of 5 × 104 cells were plated in each well of a 6-well plate. Once the confluence had reached >90%, a 200 μL pipette tip was used to scratch five wounds in the cell layer. PBS was used to gently remove floating cells, and serum-free medium containing was added to each well. The wounds were imaged at 48 h after scratching.
Transwell Assay
The migrated cells were detected by transwell assay using a 24-Transwell plate (8-µm pore size; Corning Inc., Corning, Toledo, NY, USA). Briefly, cells (5.0 × 104) in medium supplemented with 1% FBS were plated into the upper chamber non-precoated membrane. The medium containing 15% FBS was added into the lower chamber as a chemoattractant. After culture for 24 h, the migrated cells were fixed, stained, and counted under a light microscope (Olympus, Tokyo, Japan).
Cell Cycle
Cells were fixed with ethanol and then incubated with propidium iodide (PI; Solarbio, Beijing, China) for 25 min. Cells at different phases were measured with a flow cytometer (Becton Dickinson, San Jose, CA, USA).
Caspase-3 Activity
The two cells of each group were incubated in lysis buffer. After the centrifugation at 600 g for 5 min, the supernatant was collected for caspase-3 activity analysis using a caspase 3 assay kit (Beyotime, Shanghai, China) according to the instructions of the manufacturer.
Cell Apoptosis
Apoptosis rate of SW872 and 93T449 cells was assessed by using Annexin V-FITC/PI Assay Kit (Sigma), following the producer’s instructions. With Annexin V-FITC/PI double-labeling, cell apoptosis was evaluated by a flow cytometer (BD Biosciences, Basel, Switzerland) with BD FACSDiva software.
Bioinformatics and Dual-Luciferase Reporter
Bioinformatic analysis of the direct interacted miRNA target sites was performed using DIANA online tool (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index). OSBP 3ʹ-UTR wild-type reporter (OSBP-WT) harboring the miR-195-5p-binding sequence was constructed using pmirGLO vector (Promega) as previously described.26 Site-directed mutants (OSBP-MUT) was constructed using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs, Frankfurt, Germany). The constructs were transfected into cells, respectively, together with mimic NC or miR-195 using DharmaFECT 4 transfection reagent. Relative luciferase activity was normalized to Renilla luciferase activity at 48 h post-transfection.
RNA Immunoprecipitation (RIP)
RIP assay was conducted applying with Magna RNA immunoprecipitation kit (Millipore, Bedford, MA, USA) following the user’s guideline. In short, harvested SW872 and 93T449 cells were incubated with magnetic beads pre-coated with antibodies against Argonaute2 (anti-Ago2; Abcam, Cambridge, United Kingdom, ab32381) using anti-Immunoglobulin G (anti-IgG; Abcam, ab205718) as the negative control. After the extraction of RNA, qRT-PCR was used for measuring RNA enrichment. Cell lysates un-incubated with magnetic beads acted as the positive control (In-put).
Western Blot
Western blot assay was carried out as previously described.27 Total protein (50 µg) was electrophoretically separated and transferred onto a nitrocellulose membrane (Millipore, Etobicoke, ON, Canada). Primary antibodies used in the study included anti-OSBP (ab126182, Abcam, Cambridge, UK) and anti-β-actin (ab95437, Abcam). The secondary antibodies included horseradish peroxidase-conjugated goat anti-rabbit IgG (ab6721, Abcam). Protein bands were determined using chemiluminescence Plus Detection reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and the Image Reader LAS-4000 (Fuji Ltd, Osaka, Japan).
In vivo Assay
The nude mice were obtained from Henan Research Center of Laboratory Animal (Zhengzhou, Henan, China). Approximately 2.0×106 SW872 cells stably transfected with miR-195 or miR-con were subcutaneously injected into the nude mice to generate xenograft in vivo (n=8 each group). After 7 days, tumor volume was detected with a digital caliper every 7 days. All mice were euthanized after 35 days, and tumor weight was measured. Our study was approved by the Animal Care and Use Committee of the Shanghai Public Health Clinical Center, Fudan University.
Statistical Analysis
Comparison of groups was carried out using a Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test, which was performed by GraphPad Prism 7.0 software (GraphPad Software Inc., San Diego, CA, USA). The Spearman test was performed to evaluate the correlation between miR-195 and OSBP in liposarcoma tissues. Error bars represented the standard deviations. Statistical significance was set as P < 0.05.
Results
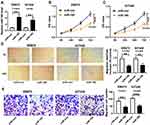
miR-195 Was Decreased in Liposarcoma
We detected the expression of miR-195 in 28 liposarcoma tissues from patients and 8 normal fat tissues from volunteers by qRT-PCR. The result showed that miR-195 expression was significantly lower in liposarcoma tissues than that in normal fat tissues (Figure 1A). The low level of miR-195 was related to the poor overall survival rate (Figure 1B). Overall, miR-195 might be an indicator of liposarcoma diagnosis.
miR-195 Overexpression Suppressed Proliferation, Migration and Induced Cell Apoptosis in SW872 and 93T449 Cells
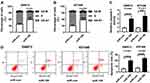
As shown in Figure 2A, qRT-PCR assay confirmed a successful overexpression efficiency of miR-195 in SW872 and 93T449 cells. Moreover, miR-195 upregulation significantly inhibited cell proliferation in SW872 and 93T449 cells (Figure 2B and C). Wound healing and transwell assays indicated that overexpression of miR-195 suppressed cell migration in SW872 and 93T449 cells (Figure 2D and E). Besides, miR-195 overexpression increased the G0-G1 phase cells (40% ~ 60%) (Figure 3A and B). The caspase-3 activity was dramatically upregulated by miR-195 overexpression in SW872 and 93T449 cells (Figure 3C). Flow cytometry analysis indicated that miR-195 overexpression promoted cell apoptosis (Figure 3D). These results showed that miR-195 overexpression suppressed cell proliferation, migration and elevated cell apoptosis in SW872 and 93T449 cells.
miR-195 Directly Targeted OSBP
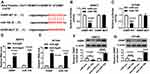
To further explore the regulatory mechanism of miR-195, we speculated that miR-195 may have a putative binding site with ZEB1-AS1 by DIANA tool (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) (Figure 4A). Then, a dual-luciferase reporter assay was performed and the results indicated that the luciferase activity of pMIR-REPOR-OSBP-WT was reduced by miR-195, but the luciferase activity was not changed in SW872 and 93T449 cells co-transfected with pMIR-REPOR-OSBP-MUT and miR-195 (Figure 4B and C). RIP assay further confirmed the direct interaction between miR-195 and OSBP, and a significant amount of miR-195 and OSBP was detected in Anti-Ago2 compared with negative Anti-IgG control (Figure 4D and E). Moreover, OSBP protein level could be restrained by miR-195 overexpression and elevated miR-195 deletion (Figure 4F and G). Taken together, miR-195 directly interacted with OSBP.
OSBP Was Increased in Liposarcoma
As shown in Figure 5A and B, qRT-PCR and Western blot showed that the mRNA and protein expression of OSBP were enhanced in liposarcoma tissues (Figure 5A and B). Moreover, OSBP expression was negatively correlated with miR-195 level (Figure 5C).
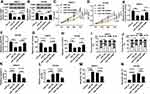
OSBP Rescued the Effects of miR-195 on Cell Proliferation, Migration and Apoptosis in SW872 and 93T449 Cells
Then, miR-con, miR-195, miR-195+pcDNA, or miR-195+OSBP were transfected into SW872 and 93T449 cells. Western blot exhibited that OSBP overexpression restored the expression of OSBP weakened by miR-195 (Figure 6A and B). Moreover, the inhibition effect of miR-195 on cell proliferation was reversed by OSBP upregulation (Figure 6C and D). Consistently, OSBP overexpression could partially restore the suppressive effect of miR-195 upregulation on cell migration (Figure 6E–H). In terms of miR-195 administrated-cell cycle and apoptosis, OSBP successfully recovered G0-G1 phase arrest (Figure 6I and J), inverted promoted-apoptotic cells (Figure 6M and N) as well as enhanced-caspase-3 activity (Figure 6K and L). To sum up, OSBP could rescue the effects of miR-195 on cell proliferation, migration and apoptosis in SW872 and 93T449 cells.
miR-195 Overexpression Inhibited Tumor Growth in Liposarcoma in vivo
Then, we further investigated the effect of miR-195 overexpression on tumor growth in xenograft mice in vivo. About 2.0×106 SW872 cells transfected with miR-con or miR-195 were subcutaneously injected into the nude mice. In contrast to the negative control, miR-195 overexpression significantly suppressed tumor growth, as evidenced by a decrease of tumor volume and average weight (Figure 7A and B). Moreover, miR-195 expression was markedly increased, and OSBP protein expression was strikingly decreased in tumor tissues derived from miR-195-transfected SW872 cells (Figure 7C and D). These data suggested that miR-195 overexpression might inhibit tumor growth in vivo by sponging OSBP.
Discussion
A previous paper studied by Li et al28 revealed that miR-195 expression was suppressed in colon carcinoma tissues and negatively correlated with the metastatic potential. Besides, miR-195 decreased the proliferative, migrative/invasive capacities of colon cancer cells, while miR-195 inhibition exerted opposing effects. miR-195 synergized with microtubule-targeting agents (MTAs) inhibited the non-small cell lung cancer cell growth in vitro, while increased miR-195 expression sensitized NSCLC cells to MTAs and miR-195 repression conferred resistance to MTAs.29 Consistent with Li et al paper, we initially examined the expression of miR-195 in liposarcoma tissues from patients, qRT-PCR detection showed that miR-195 was decreased in tissues of liposarcoma as well as its suppressed-expression could predict a poor overall survival (%).
To further evaluate the bio-function effects in vitro, we measured the effects of miR-195 overexpression on proliferation, migration, cell cycle arrest, and apoptosis, the results confirmed that miR-195 upregulation suppressed proliferation and migration, and induced cell growth inhibition and apoptosis in SW872 and 93T449 cells. These data manifested that miR-195 founctioned as an essential miRNA to regulate the progression of liposarcoma in vitro. Moreover, miR-195 overexpression significantly repressed tumor growth in vivo.
DIANA software was an online tool, which could be used to explore downstream genes. As shown in the part of the research, we tracked that OSBP was a target gene for miR-195. The dual-luciferase reporter gene assay and RIP assay verified the interrelationship between miR-195 and OSBP. Besides, Western blot results indicated that miR-195 negatively regulated the OSBP level in both AW872 and 93T449 cells.
Emerging evidence have demonstrated that OSBP promoted cancer development. Suppression of OSBP significantly inhibited cell growth and AKT phosphorylation in SW872-S cells.25 In our experiments, enhanced levels of OSBP, both at mRNA and protein levels, were observed in the liposarcoma tissues. Meanwhile, a negative correlation between miR-195 and OSBP was found in liposarcoma tissues. In the subsequent bio-functional assays, we overexpressed OSBP expression to examine whether it could rescue the effect caused by miR-195. As we expected, OSBP successfully inverted the inhibition on proliferation and migration and rescued the promotion on apoptosis and cell cycle arrest (G0-G1) caused by miR-195 upregulation. However, the study limitation in this research was that commercial cell lines we selected could not represent all kinds of liposarcoma cells.
Conclusion
We concluded that miR-195 suppressed cell proliferation, migration and enhanced cell apoptosis via OSBP in liposarcoma.
Abbreviations
OSBP, oxysterol-binding protein; RIP, RNA immunoprecipitation.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Mack TM. Sarcomas and other malignancies of soft tissue, retroperitoneum, peritoneum, pleura, heart, mediastinum, and spleen. Cancer. 1995;75(1 Suppl):211–244. doi:10.1002/1097-0142(19950101)75:1+<211::aid-cncr2820751309>3.0.co;2-x
2. Conyers R, Young S, Thomas DM. Liposarcoma: molecular genetics and therapeutics. Sarcoma. 2011;2011:483154. doi:10.1155/2011/483154
3. Crago AM, Singer S. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol. 2011;23(4):373–378. doi:10.1097/CCO.0b013e32834796e6
4. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
5. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
6. Guo H, Ingolia NT, Weissman JS, et al. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi:10.1038/nature09267
7. Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi:10.1101/gr.082701.108
8. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. doi:10.1038/nrg2634
9. Ventura A, Jacks T. MicroRNAs and cancer: short RNAs go a long way. Cell. 2009;136(4):586–591. doi:10.1016/j.cell.2009.02.005
10. Li M, Lee KF, Lu Y, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer Cell. 2009;16(6):533–546. doi:10.1016/j.ccr.2009.10.025
11. Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438(7068):685–689. doi:10.1038/nature04303
12. Casadei L, Calore F, Creighton CJ, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 2017;77(14):3846–3856. doi:10.1158/0008-5472.CAN-16-2984
13. Yu PY, Lopez G, Braggio D, et al. miR-133a function in the pathogenesis of dedifferentiated liposarcoma. Cancer Cell Int. 2018;18:89. doi:10.1186/s12935-018-0583-2
14. Hu WY, Wei HY, Li KM, et al. LINC00511 as a ceRNA promotes cell malignant behaviors and correlates with prognosis of hepatocellular carcinoma patients by modulating miR-195/EYA1 axis. Biomed Pharmacother. 2019;121:109642. doi:10.1016/j.biopha.2019.109642
15. Ma S, Chen C, Ji X, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol. 2019;12(1):121. doi:10.1186/s13045-019-0805-7
16. Liang Y, Rong X, Luo Y, et al. A novel long non-coding RNA LINC00355 promotes proliferation of lung adenocarcinoma cells by down-regulating miR-195 and up-regulating the expression of CCNE1. Cell Signal. 2019;66:109462. doi:10.1016/j.cellsig.2019.109462
17. Yu Y, Dong JT, He B, et al. LncRNA SNHG16 induces the SREBP2 to promote lipogenesis and enhance the progression of pancreatic cancer. Future Oncol. 2019;15(33):3831–3844. doi:10.2217/fon-2019-0321
18. Chae DK, Park J, Cho M, et al. MiR-195 and miR-497 suppress tumorigenesis in lung cancer by inhibiting SMURF2-induced TGF-beta receptor I ubiquitination. Mol Oncol. 2019;13(12):2663–2678. doi:10.1002/1878-0261.12581
19. Fricke A, Ullrich PV, Heinz J, et al. Identification of a blood-borne miRNA signature of synovial sarcoma. Mol Cancer. 2015;14:151. doi:10.1186/s12943-015-0424-z
20. Pietrangelo A, Ridgway ND. Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell Mol Life Sci. 2018;75(17):3079–3098. doi:10.1007/s00018-018-2795-y
21. Brown MS, Dana SE, Goldstein JL. Cholesterol ester formation in cultured human fibroblasts. Stimulation by oxygenated sterols. J Biol Chem. 1975;250(10):4025–4027.
22. Taylor FR, Kandutsch AA. Oxysterol binding protein. Chem Phys Lipids. 1985;38(1–2):187–194. doi:10.1016/0009-3084(85)90066-0
23. Du X, Turner N, Yang H. The role of oxysterol-binding protein and its related proteins in cancer. Semin Cell Dev Biol. 2018;81:149–153. doi:10.1016/j.semcdb.2017.07.017
24. Im YJ, Raychaudhuri S, Prinz WA, et al. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437(7055):154–158. doi:10.1038/nature03923
25. Chen W, Qian X, Hu Y, et al. SBF-1 preferentially inhibits growth of highly malignant human liposarcoma cells. J Pharmacol Sci. 2018;138(4):271–278. doi:10.1016/j.jphs.2018.10.009
26. Liu C, Pan C, Cai Y, et al. Interplay between long noncoding RNA ZEB1-AS1 and miR-200s regulates osteosarcoma cell proliferation and migration. J Cell Biochem. 2017;118(8):2250–2260. doi:10.1002/jcb.25879
27. Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol. 2019;862:172615. doi:10.1016/j.ejphar.2019.172615
28. Li B, Wang S, Wang S. MiR-195 suppresses colon cancer proliferation and metastasis by targeting WNT3A. Mol Genet Genomics. 2018;293(5):1245–1253. doi:10.1007/s00438-018-1457-y
29. Yu X, Zhang Y, Ma X, et al. miR-195 potentiates the efficacy of microtubule-targeting agents in non-small cell lung cancer. Cancer Lett. 2018;427:85–93. doi:10.1016/j.canlet.2018.04.007
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.