Back to Journals » Cancer Management and Research » Volume 11
MiR-193a-3p inhibits pancreatic ductal adenocarcinoma cell proliferation by targeting CCND1
Authors Chen ZM, Yu Q, Chen G , Tang RX, Luo DZ, Dang YW, Wei DM
Received 22 December 2018
Accepted for publication 29 March 2019
Published 27 May 2019 Volume 2019:11 Pages 4825—4837
DOI https://doi.org/10.2147/CMAR.S199257
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Zhi-Min Chen,* Qiao Yu,* Gang Chen, Rui-Xue Tang, Dian-Zhong Luo, Yi-Wu Dang, Dan-Ming Wei
Department of Pathology, First Affiliated Hospital of Guangxi Medical University, Nanning, People’s Republic of China
*These authors contributed equally to this work
Background: MicroRNAs (miRNAs) could modulate gene expression at the posttranscriptional level by promoting mRNA degradation or blocking mRNA translation, thus affecting the occurrence and development of cancer.
Methods: In this work, qRT-PCR was conducted to detect the expression of miR-193a-3p and CCND1. The ability of cell proliferation was evaluated via CCK-8 assay. Cell apoptosis and cell cycle distribution were detected by flow cytometry. Bioinformatic techniques were employed to research the regulatory relationship between miR-193a-3p and target genes. The relationship between miR-193a-3p and CCND1 was verified via dual-luciferase reporter assays.
Results: MiR-193a-3p expression in pancreatic ductal adenocarcinoma (PDAC) tissue was significantly lower than in non-cancerous tissue. After overexpressing miR-193a-3p in PDAC cells, their multiplication ability was significantly inhibited, apoptosis was accelerated, and the cell cycle was blocked in the G1 and G2/M phases. CCND1 was confirmed to have a targeted relationship with miR-193a-3p. Moreover, CCND1 expression was significantly lower in PDAC cells with an overexpression of miR-193a-3p.
Conclusions: MiR-193a-3p targeted CCND1 to suppress tumor growth in PDAC cells. MiR-193a-3p may function as a tumor inhibitor in PDAC development, which could offer a promising therapeutic and prognostic strategy for PDAC treatment.
Keywords: miR-193a-3p, CCND1, PDAC, proliferation
Introduction
Pancreatic ductal adenocarcinoma (PDAC) accounts for 90 percent of pancreatic cancer, with a 5-year survival rate under 5 percent.1 It is primarily caused by the biological characteristics of early recurrence and metastasis and the lack of early diagnosis methods.2,3 MicroRNAs (miRNAs), a sort of noncoding RNA, have approximately 22 ribonucleotide RNA sequences.3 MiRNAs are very stable in tissue, plasma, feces, and other liquids, and can be quantitatively analyzed using a very small sample size for clinical detection.4 Regarded as either oncogenes or anti-oncogenes, miRNAs help regulate posttranscriptional gene expression.5 Several works have indicated that miRNAs can be identified as biomarkers to diagnose and evaluate the prognosis of PDAC.6,7 For instance, PDAC patients with miR-1-positive tumors had a longer survival time, and miR-1 status was associated with tumor size and TMN stage.8
MiR-193a-3p was first reported by Lagos-Quintana et al, who found that it was a tumor inhibitor and transforming factor of parietal cells in 2003.9 It has now been discovered to play a vital role in multiple diseases. In acute myeloid leukemia, miRNA-193a-3p acts as a tumor suppressor to repress the expression of KRAS.10 Other studies show that miR-193a-3p participates in inhibiting tumor progression in lung cancer11 and colorectal cancer.12
Here, we researched the effect of miR-193a-3p on the proliferation and invasion of PDAC cells and looked into the correlation between miR-193a-3p and the target gene CCND1 using prediction software as well as experimental verification. We found that miR-193a-3p could act as a tumor inhibitor in PDAC by directly targeting CCND1.
Material and methods
Tissue samples
We collected 37 samples of formalin-fixed paraffin-embedded (FFPE) PDAC tissue and 42 samples of non-cancerous pancreas tissue from the pathology department of the First Affiliated Hospital of Guangxi Medical University. These samples were confirmed by pathology to come from patients between January 2012 and June 2017 who were treated with neither radiation nor chemotherapy before resection. Written informed consents were obtained from the patients before surgery. This study was approved by the Ethics Committee of the First Hospital of Guangxi Medical University, and conducted in accordance with the Declaration of Helsinki.
Cell culture
BxPC-3, PANC-1, and Hpde6-C7 cell lines were all accessed from the Cell Bank, Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). These cells were artificially nurtured in a proportionally prepared medium including Dulbecco’s modified Eagle’s medium, RPMI-1640 medium, fetal bovine serum, and penicillin/streptomycin (Gibco, Grand Island, NY, USA). Culture bottles were put in a humidified incubator with 5% CO2.
qRT‑PCR
For tissues and cells, all the RNA was extracted according to the instructions of the E.Z.N.A.® FFPE RNA Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) and Total RNA Kit I (Omega). Subsequently, the complementary DNAs (cDNAs) were inversely transcribed in line with the Transcriptor First Strand cDNA Synthesis Kit (Roche, Mannheim, Germany) or the miRNA First Strand cDNA Synthesis (Tailing Reaction) (Sangon Biotech, Shanghai, China). The expression levels of miRNA relative to U6 and those of mRNA relative to were accessed via qRT‑PCR, which was conducted by employing the FastStart Universal SYBR-Green Master (Roche) or the MicroRNAs qPCR Kit (SYBR Green Method) (Sangon Biotech) on an ABI Prism 7500 (Applied Biosystems, Foster City, CA, USA).
Transfection of lentivirus
The overexpression of miR-193a-3p was conducted via transfecting lentivirus. The PDAC cell suspension was seeded into 6-well plates (2 ml/wall) and then transfected by lentivirus for 16 h, followed by changing the transfection medium into a routine medium. The group with the overexpression of miR-193a-3p was the OE group.
CCK-8 assay
Transfected cells were planted in 96-well plates (2000 cells/well) for 24, 48, 72, 96, or 120 h. The CCK-8 reagents were added to each well three hours before the end of culture. A microplate reader was employed to determine the absorbance at 450 nm.
PI-FACS analysis
Transfected cells were washed and collected with precooled D-Hanks (pH=7.2~7.4). After centrifugation, precooled 75% ethanol was used to fix the cells for 1 h. According to the cell quantity, a PI solution was added to re-suspend the cells, so that the cell passing rate ranged from 300~800 cells/s when the cells were on the flow cytometer. Lastly, the cell cycle distribution was observed.
Apoptosis analysis
Using the method of Annexin V-APC Single Staining, we used 4 °C pre-cooled D-Hanks, PBS(phosphate buffered saline), and a 1×binding buffer to wash the cell pellet once (1500 rpm, 5 min centrifuged); then the 1×staining buffer was used to re-suspend the cell precipitation and Annexin V-APC was added to stain it for 10–15 min, keeping it in a dark place at room temperature before conducting flow cytometry.
Dual-luciferase reporter assays
Luciferase activity was identified using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA). HEK293T cells were transfected with the target gene 3‘ UTR plasmids (CCND1 3‘ UTR+miR-193a-3p) or no-load plasmids (CCND1 3‘ UTR-NC+miR-193a-3p). To make the experimental results more reliable, both the positive reference miRNA group (has-miR-146b vector plasmid) and the positive reference miRNA NC(negative control) group (TRAF6 3′UTR plasmids) were set up.
Bioinformatics analysis
The miR-193a-3p target genes were predicted using the target gene prediction software miRwalk (
Datasets related to miR-193a-3p and CCND1 expression were searched for in the databases of GEO (Gene Expression Omnibus) using the search strategy. We searched using the following search strategy: “(pancreas OR pancreatic) and (tumor OR cancer OR carcinoma OR adenocarcinoma OR malignan* OR neoplas* OR PDAC OR PC OR PAAD)”. The expression of miR-193a-3p was also obtained from online database (
Results
miR-193a-3p expression in PDAC
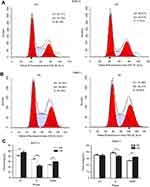
We found the expression of miR-193a-3p was down-regulation in PDAC by analyzing data from online database (Figure 1A). The result of qRT-PCR suggested that miR-193a-3p expression was decreased in the PDAC cell lines (Figure 1B). In addition, the expression levels were verified in the 37 cases of PDAC comparing with the 42 cases of adjacent pancreatic tissues. Likewise, miR-193a-3p expression in PDAC tissue was significantly lower than that in tumor adjacent tissue (Figure 1C). Moreover, combing with database mining, there was a low expression of miR-193a-3p in PDAC patients (Figure 1D).
Effect of miR-193a-3p overexpression on PDAC cells
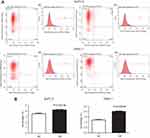
The results of the CCK-8 assay indicated that cell proliferation could be repressed by increasing miR-193a-3p expression in PDAC cell lines (Figure 2). Compared to the NC group, the proliferation capacity of BxPC-3 cells was significantly decreased 3 d(P=0.001), 4 d(P=0.022), and 5 d(P=0.002) after transfection, while the multiplication ability of Panc-1 cells was significantly reduced 2 d(P=0.006), 3 d(P=0.001), 4 d(P=0.011), and 5 d(P=0.000) after transfection. Afterwards, we carried out a flow cytometry analysis to investigate whether miR-193a-3p impacts PDAC cell proliferation by altering the cell cycle distribution. Compared with the control group, transfected PDAC cells were both visibly blocked in the G2/M phase, while cells in the S phase were reduced. BxPC-3 cells that were overexpressed with miR-193a-3p also had cell cycle arrest in the G1 phase (Figure 3). According to the results of the Annexin V-APC Single Staining, the overexpression of miR-193-3p significantly induced BxPC-3 and Panc-1 cell apoptosis (Figure 4).
Target gene analysis of miR-193a-3p
The 285 genes that are repeatedly detected by at least eight platforms were selected for later works. The GO analysis showed that these genes were concentrated in 71 pathways, 9 of which were significant (P≤0.01; Figure 5, Table 1). The KEGG analysis of these 285 genes resulted in 37 signaling pathways, of which 14 were significant (P≤0.01; Figure 5, Table 2). Interestingly, the gene CNND1 functioned in many tumor-related pathways, such as Pathways in cancer and MicroRNAs in cancer. What is more, the results of the protein-protein interaction network analysis showed that CCND1 had a high interaction with all the pivotal genes (Figure 6).
 | Table 1 GO analysis of miR-193a-3p target genes |
 | Table 2 KEGG pathway analysis of miR-193a-3p target genes |
 | Figure 6 Protein-protein interaction network of miR-193a-3p target genes. |
CCND1 expression in PDAC
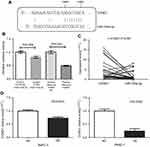
Our qRT-PCR data illustrated that CCND1 expression in FFPE PDAC tissues (3.2482±4.3472) was significantly higher than in non-tumor tissues (1.5356±1.64881; Figure 7A). Meanwhile, we mined 11 databases to detect CCND1 expression in PDAC and para-carcinoma pancreatic tissues by searching GEO. Subsequently, a forest plot of CCND1 expression was performed by combining our data with the published findings. The results illustrated that CCND1 expression in PDAC tissues was significantly higher than that of normal pancreatic tissues (SMD=0.69, CI=0.38–0.99, P<0.001; Figure 7B).
Correlation between miR-193a-3p and CCND1
Through searching online prediction software, we found that there were complementary bases between miR-193a-3p and CCND1 (Figure 8A). The results of the dual-luciferase reporter assay showed that there was direct binding sites between miR-193a-3p and CCND1 (Figure 8B). The expression of miR-193a-3p and CCND1 were detected in the same samples. After matching the samples, a correlation analysis was performed. The result showed the correlation was not statistically significant (r=−0.0021, P=0.99; Figure 8C). More notably, CCND1 expression was significantly decreased in PDAC cells with miR-193a-3p overexpression (Figure 8D).
Discussion
The occurrence and development of PDAC is a complex, multistep process involving multiple genes and epigenetic changes.13 It is needed to investigate the molecular basis of the occurrence and development of PDAC in order to seek more effective diagnostic and therapeutic methods. Over the past decades, numerous studies have demonstrated the important role of miRNAs in the progression of multiple tumors. MiRNAs control various tumor-related courses, including propagation, mobility and invasion, apoptosis, angiogenesis, and even the transdifferentiation of stem cells.14,15 MiRNA is considered to be not only a biomarker and prognostic factor, but also a potential treatment for certain cancers.16
MiR-193a-3p, a member of the miR-193 family, is located at 17q11.2. Recently, it was reported that miR-193a-3p expression was decreased in tumors such as colorectal cancer,17 lung cancer,10 and liver cancer.18 Likewise, we found that miR-193a-3p expression in 37 cases of PDAC tissues was significant lower than in the 42 cases of non-tumor tissues. The low level of miR-193a-3p expression was examined in PANC-1 and BxPC-3. It also turned out to be down-regulated in PDAC after we performed a meta-analysis combining the qRT-PCR experiment with data from GEO. The down-regulated expression of miR-193a-3p corresponded to a poor prognosis.12
Recent studies have uncovered that miR-193a-3p was involved in several biological processes such as proliferation, apoptosis, and migration.19,20 We researched the biological functions of PANC-1 and BxPC-3 to investigate how miR-193a influences the biological behavior of PDAC cells. In this work, after overexpressing miR-193a-3p in PDAC cells, we found the cell-cycle was blocked in the G1 and G2/M phases. Although the apoptosis analysis showed that the cell apoptosis rate was low (<5%), the overexpression miR-193a-3p had a tendency to promote pancreatic cancer cell apoptosis and the difference was statistically significant. The results were consistent with the results that the cell proliferation was significantly inhibited after the overexpression of miR-193a-3p in the CCK-8 assay. Existing research has demonstrated that miRNAs could influence the occurrence of colorectal cancer through cell cycle control.21,22 Colon cancer cells were blocked during the transition from S phase to G2/M phase; however, cells were detained in the G2/M phase when miR-193a was overexpressed in the cells, thereby promoting the apoptosis of the colon cells.23 The transition from the G1 and S phases was synergistically inhibited by miR-193a-3p and other miRNAs in breast cancer.24 In addition, miR-193a-3p inhibited breast cancer cell proliferation and apoptosis by inhibiting the expression of PTP1B.25 Thus, miR-193a-3p expression was lower in PDAC and an overexpression of miR-193a-3p could repress cell proliferation and motivate the apoptosis of PDAC cells, which indicates that miR-193a-3p may be a potential tumor inhibitor for PDAC.
At present, there are a number of studies that indicated miR-193a-3p influences tumor progression by targeting genes. MiR-193a-3p down-regulation could inhibit cell proliferation, migration, and chemical resistance by targeting PTEN(phosphatase and tensin homolog)in stomach cancer26 and could inhibit cell multiplication and migration in renal cell cancer.27 LINC00152 has been shown to enhance gastric cancer cell proliferation by down-regulating miR-193a-3p expression and increasing MCL1 expression.28 MiR-193a-3p took part in regulating the proliferation by targeting c-kit in AML cells29 and by affecting the expression of ERBB4 in lung cancer cells.30 MiR-193a-3p also inhibited cell proliferation metastasis in renal cancer by targeting PTEN27 and induced apoptosis by targeting ST3GalIV.31
In this study, we applied target gene prediction tools to explore whether miR-193a-3p suppresses PDAC cell proliferation by targeting genes. As a result, CCND1 was found to be key gene in the network analysis and was concentrated in a variety of tumor pathways. The CCND1 gene, located on chromosome 11q13, was overexpressed in multiple human cancers of the breast,32 stomach,33 and ovarian cancer34 as a well-known oncogene. In pancreatic cancer, CCND1 expression was high and significantly associated with the degree of differentiation and a poor prognosis.35 A functional analysis showed that a CCND1 knockdown could inhibit cell proliferation, soft AGAR cloning, metastasis, metabolism, and cell growth.36,37 The reduced expression of CCND1 enhanced the chemotherapy sensitivity of patients to 5-fluorouracil, 5-fluoro-2‘-deoxyuridine, and mitoxantrone by down-regulating various chemo-resistant genes in pancreatic cancer.38 Therefore, the regulation of the miR-193a-3p/CCND1 axis may be a new method of treatment to repress the fast growth and metastasis of pancreatic cancer.
We explored the correlation between CCND1 and miR-193a-3p and found there were bases’ binding sites between them. qRT-PCR detection in pancreatic cancer cells with an overexpression of miR-193a-3p indicated that CCND1 expression significantly decreased compared with the control group. This result suggested that miR-193a-3p can negatively regulate CCND1 expression at the mRNA level in PDAC cells. Furthermore, miR-193a-3p overexpression reduced the luciferase activity of CCND1 3ʹUTR, which demonstrated that a direct binding site existed between miR-193a-3p and CCND1. However, there was no significantly negative correlation in their expression (R=−0.0021). Perhaps the insignificantly statistic difference of negative correlation resulted from the deficient number of cases. In normal physiological functions, CCND1 acted as a key sensor, mediating cellular functions by binding to cyclin-dependent kinases. CCND1 could make the function of suppressing the cell cycle of retinoblastoma proteins inactive as well.39 Therefore, miR-193a-3p overexpression led to the PDAC cell cycle’s block at the G1 phase, which have been caused by inhibiting CCND1 expression. Nevertheless, CCND1 may have other carcinogenic functions besides its influence on the cell cycle. Studies on tumor models revealed that CCND1 could act as a transcriptional regulatory factor and potentially cause chromosomal instability.40,41 It is worth noting that previously CCND1 was discovered to promote DNA repair by binding with recombinant activator genes and homologous DNA recombination,42 which was also considered to be the cause of anti-apoptosis of cancer cells. In this study, CCND1 expression was decreased after miR-193a-3p overexpression in PDAC cells. The presence of a base binding site between miR-193a-3p and CCND1 was detected by bioinformatics prediction software. Beyond that, the luciferase assay showed that the former inhibited the latter’s expression by binding directly to 3ʹUTR at the post-transcriptional level. Therefore, the inhibition of CCND1 expression in pancreatic cancer may be a promising therapeutic strategy.
Conclusion
Our findings illustrated a potential tumor suppressor function of miR-193a-3p in PDAC progress by targeting CCND1, which offers new ideas for exploring the molecular mechanism of PDAC and may provide a possible therapeutic strategy for PDAC treatment. However, the mechanism of their interaction in pancreatic cancer has not been fully elucidated and further studies are needed in deeper experiments.
Abbreviation list
PDAC, pancreatic ductal adenocarcinoma; miRNAs, microRNAs; FFPE, formalin-fixed paraffin-embedded; cDNAs, complementary DNAs; PBS, Phosphate Buffered Saline; GO,gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GEO, Gene Expression Omnibus; AML, acute myeloid leukemia; PTEN, phosphatase and tensin homolog.
Acknowledgments
The study was supported by the funds of the National Natural Science Foundation of China (NSFC81560448, NSFC81560489), the Natural Science Foundation of Guangxi, China (2014GXNSFBA118167, 2016GXNSFBA380039, 2018GXNSFAA050051), Guangxi Medical University Training Program for Distinguished Young Scholars (2017), and the Promoting Project of Basic Capacity for University Young and Middle-aged Teachers in Guangxi (KY2016LX031). This study also received a Medical Excellence Award, funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University, and was also a Guangxi Zhuang Autonomous Region Health and Family Planning Commission Self-financed Scientific Research Project (Z20180979). The authors appreciate all the publicly available data used in the current study. The funders had no role in the study design, the data collection and analysis, the decision to publish, or the preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Song HY, Wang Y, Lan H, Zhang YX. Expression of Notch receptors and their ligands in pancreatic ductal adenocarcinoma. Exp Ther Med. 2018;16(1):53–60. doi:10.3892/etm.2018.6172
2. You L, Wang J, Zhang F, et al. Potential four-miRNA signature associated with T stage and prognosis of patients with pancreatic ductal adenocarcinoma identified by co-expression analysis. Mol Med Rep. 2019;19(1):441–451. doi:10.3892/mmr.2018.9663
3. Chang RK, Li X, Mu N, et al. MicroRNA expression profiles in nonepithelial ovarian tumors. Int J Oncol. 2018;52(1):55–66.
4. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
5. Wei DM, Dang YW, Feng ZB, et al. Biological effect and mechanism of the miR-23b-3p/ANXA2 axis in pancreatic ductal adenocarcinoma. Cell Physiol Biochem. 2018;50(3):823–840. doi:10.1159/000494468
6. D‘Angelo B, Benedetti E, Cimini A, Giordano A. MicroRNAs: a puzzling tool in cancer diagnostics and therapy. Anticancer Res. 2016;36(11):5571–5575. doi:10.21873/anticanres.11142
7. Huang J, Liu J, Chen-Xiao K, et al. Advance in microRNA as a potential biomarker for early detection of pancreatic cancer. Biomarker res. 2016;4:20. doi:10.1186/s40364-016-0074-3
8. Cheng Q, Han LH, Zhao HJ, Li H, Li JB. Abnormal alterations of miR-1 and miR-214 are associated with clinicopathological features and prognosis of patients with PDAC. Oncol Lett. 2017;14(4):4605–4612. doi:10.3892/ol.2017.6819
9. Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. Rna. 2003;9(2):175–179.
10. Fan Q, Hu X, Zhang H, et al. MiR-193a-3p is an important tumour suppressor in lung cancer and directly targets KRAS. Cell Physiol Biochem. 2017;44(4):1311–1324. doi:10.1159/000485491
11. Deng W, Yan M, Yu T, et al. Quantitative proteomic analysis of the metastasis-inhibitory mechanism of miR-193a-3p in non-small cell lung cancer. Cell Physiol Biochem. 2015;35(5):1677–1688. doi:10.1159/000373981
12. Lin M, Duan B, Hu J, et al. Decreased expression of miR-193a-3p is associated with poor prognosis in colorectal cancer. Oncol Lett. 2017;14(1):1061–1067. doi:10.3892/ol.2017.6266
13. Zhang Q, Zeng L, Chen Y, et al. Pancreatic cancer epidemiology, detection, and management. Gastroenterol Res Pract. 2016;2016:8962321. doi:10.1155/2016/8962321
14. Morgado AL, Rodrigues CM, Sola S. MicroRNA-145 regulates neural stem cell differentiation through the Sox2-Lin28/let-7 signaling pathway. Stem Cells. 2016;34(5):1386–1395. doi:10.1002/stem.2309
15. Ghosh A, Dasgupta D, Ghosh A, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8(3):e2706. doi:10.1038/cddis.2017.123
16. Shah MY, Ferrajoli A, Sood AK, Lopez-Berestein G, Calin GA. microRNA therapeutics in cancer - An emerging concept. EBioMedicine. 2016;12:34–42. doi:10.1016/j.ebiom.2016.09.017
17. Takahashi H, Takahashi M, Ohnuma S, et al. microRNA-193a-3p is specifically down-regulated and acts as a tumor suppressor in BRAF-mutated colorectal cancer. BMC Cancer. 2017;17(1):723. doi:10.1186/s12885-017-3739-x
18. Liu Y, Ren F, Luo Y, Rong M, Chen G, Dang Y. Down-regulation of MiR-193a-3p dictates deterioration of HCC: a clinical real-time qRT-PCR study. Med sci monit. 2015;21:2352–2360. doi:10.12659/MSM.894077
19. Seviour EG, Sehgal V, Mishra D, et al. Targeting KRas-dependent tumour growth, circulating tumour cells and metastasis in vivo by clinically significant miR-193a-3p. Oncogene. 2017;36(10):1339–1350. doi:10.1038/onc.2016.308
20. Jia L, Gui B, Zheng D, et al. Androgen receptor-regulated miRNA-193a-3p targets AJUBA to promote prostate cancer cell migration. Prostate. 2017;77(9):1000–1011. doi:10.1002/pros.23356
21. Wang ZZ, Yang J, Jiang BH, et al. KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int J Oncol. 2018;53(5):1939–1952. doi:10.3892/ijo.2018.4546
22. Gopalan V, Smith RA, Lam AK. Downregulation of microRNA-498 in colorectal cancers and its cellular effects. Exp Cell Res. 2015;330(2):423–428. doi:10.1016/j.yexcr.2014.08.006
23. Mamoori A, Wahab R, Islam F, et al. Clinical and biological significance of miR-193a-3p targeted KRAS in colorectal cancer pathogenesis. Hum Pathol. 2018;71:145–156. doi:10.1016/j.humpath.2017.10.024
24. Uhlmann S, Mannsperger H, Zhang JD, et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol Syst Biol. 2012;8:570. doi:10.1038/msb.2011.100
25. Yu M, Liu Z, Liu Y, et al. PTP1B markedly promotes breast cancer progression and is regulated by miR-193a-3p. FEBS J. 2018;doi:10.1111/febs.14724.
26. Jian B, Li Z, Xiao D, He G, Bai L, Yang Q. Downregulation of microRNA-193-3p inhibits tumor proliferation migration and chemoresistance in human gastric cancer by regulating PTEN gene. Tumour Biol. 2016;37(7):8941–8949. doi:10.1007/s13277-015-4727-x
27. Liu L, Li Y, Liu S, et al. Downregulation of miR-193a-3p inhibits cell growth and migration in renal cell carcinoma by targeting PTEN. Tumour Biol. 2017;39(6):1010428317711951. doi:10.1177/1010428317711951
28. Huang Y, Luo H, Li F, et al. LINC00152 down-regulated miR-193a-3p to enhance MCL1 expression and promote gastric cancer cells proliferation. Biosci Rep. 2018;38:3. doi:10.1042/BSR20171607
29. Gao XN, Lin J, Li YH, et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30(31):3416–3428. doi:10.1038/onc.2011.62
30. Liang H, Liu M, Yan X, et al. miR-193a-3p functions as a tumor suppressor in lung cancer by down-regulating ERBB4. J Biol Chem. 2015;290(2):926–940. doi:10.1074/jbc.M114.621409
31. Pan Y, Hu J, Ma J, et al. MiR-193a-3p and miR-224 mediate renal cell carcinoma progression by targeting alpha-2,3-sialyltransferase IV and the phosphatidylinositol 3 kinase/Akt pathway. Mol Carcinog. 2018;57(8):1067–1077. doi:10.1002/mc.22826
32. Li J, Wei J, Mei Z, et al. Suppressing role of miR-520a-3p in breast cancer through CCND1 and CD44. Am J Transl Res. 2017;9(1):146–154.
33. Kumari S, Prasad SB, Yadav SS, et al. Cyclin D1 and cyclin E2 are differentially expressed in gastric cancer. Med oncol. 2016;33(5):40. doi:10.1007/s12032-016-0754-8
34. Dai J, Wei RJ, Li R, Feng JB, Yu YL, Liu PS. A study of CCND1 with epithelial ovarian cancer cell proliferation and apoptosis. Eur Rev Med Pharmacol Sci. 2016;20(20):4230–4235.
35. Rozengurt E, Sinnett-Smith J, Eibl G. Yes-associated protein (YAP) in pancreatic cancer: at the epicenter of a targetable signaling network associated with patient survival. Signal Transduction Targeted Ther. 2018;3:11. doi:10.1038/s41392-017-0005-2
36. Yan L, Wang Y, Wang ZZ, et al. Cell motility and spreading promoted by CEACAM6 through cyclin D1/CDK4 in human pancreatic carcinoma. Oncol Rep. 2016;35(1):418–426. doi:10.3892/or.2015.4338
37. Lee Y, Ko D, Min HJ, et al. TMPRSS4 induces invasion and proliferation of prostate cancer cells through induction of Slug and cyclin D1. Oncotarget. 2016;7(31):50315–50332. doi:10.18632/oncotarget.10382
38. Kornmann M, Danenberg KD, Arber N, Beger HG, Danenberg PV, Korc M. Inhibition of cyclin D1 expression in human pancreatic cancer cells is associated with increased chemosensitivity and decreased expression of multiple chemoresistance genes. Cancer Res. 1999;59(14):3505–3511.
39. Shi Y, Qian ZR, Zhang S, et al. Cell cycle protein expression in neuroendocrine tumors: association of CDK4/CDK6, CCND1, and phosphorylated retinoblastoma protein with proliferative index. Pancreas. 2017;46(10):1347–1353. doi:10.1097/MPA.0000000000000944
40. Vodicka P, Musak L, Vodickova L, et al. Genetic variation of acquired structural chromosomal aberrations. Mutat Res. 2018;836(Pt A):13–21. doi:10.1016/j.mrgentox.2018.05.014
41. Casimiro MC, Crosariol M, Loro E, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Invest. 2012;122(3):833–843. doi:10.1172/JCI60256
42. Jirawatnotai S, Hu Y, Michowski W, et al. A function for cyclin D1 in DNA repair uncovered by protein interactome analyses in human cancers. Nature. 2011;474(7350):230–234. doi:10.1038/nature10155
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.