Back to Journals » OncoTargets and Therapy » Volume 13
MiR-154-5p Suppresses Cell Invasion and Migration Through Inhibiting KIF14 in Nasopharyngeal Carcinoma
Authors Chen J, Ma C , Zhang Y, Pei S, Du M, Zhang Y, Qian L, Wang J, Yin L, He X
Received 20 December 2019
Accepted for publication 3 March 2020
Published 13 March 2020 Volume 2020:13 Pages 2235—2246
DOI https://doi.org/10.2147/OTT.S242939
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr William C. Cho
Jie Chen,1,2,* Chengxian Ma,2,* Yufeng Zhang,2 Shuai Pei,1,2 Mingyu Du,2 Yujie Zhang,2 Luxi Qian,2 Jianlin Wang,2 Li Yin,2 Xia He1,2
1Xuzhou Medical University, Xuzhou, Jiangsu, People’s Republic of China; 2Jiangsu Cancer Hospital, Jiangsu Institute of Cancer Research, Nanjing Medical University Affiliated Cancer Hospital, Nanjing, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xia He
Xuzhou Medical University, Xuzhou, Jiangsu, People’s Republic of China
Email [email protected]
Li Yin
Jiangsu Cancer Hospital, Jiangsu Institute of Cancer Research, Nanjing Medical University Affiliated Cancer Hospital, Nanjing, Jiangsu, People’s Republic of China
Email [email protected]
Background: Mounting evidence has reported that microRNA-154-5p (miR-154-5p) is involved in the development of multiple cancers, but its function in nasopharyngeal carcinoma (NPC) remains not well investigated.
Methods: Real-time quantitative PCR (qRT-PCR) was used to detect miR-154-5p expression in NPC tissues and cells. CCK8, colony formation, wound healing and transwell assays were performed to assess cell proliferation, migration and invasion. Dual-luciferase reporter assays and Western blots were performed to confirm the target gene of miR-154-5p. Rescue experiments were conducted to explore the influence of target gene KIF14 on the functions of miR-154-5p. Xenograft tumor model was conducted to detect the effect of miR-154-5p in vivo.
Results: qRT-PCR results revealed that the expression of miR-154-5p was down-regulated in NPC tissues and cell lines compared to normal nasopharyngeal tissues and cell line. Overexpression of miR-154-5p inhibited cell migration and invasion. However, miR-154-5p had no influence on the proliferation of NPC cells. MiR-154-5p overexpression suppressed xenograft tumor metastasis in vivo. Dual-luciferase reporter analysis identified KIF14 as a target gene of miR-154-5p. Rescue experiments showed that knockdown of KIF14 reversed the effect of inhibiting miR-154-5p expression on NPC cell migration and invasion.
Conclusion: Taken together, miR-154-5p suppresses tumor migration and invasion by targeting KIF14 in NPC. The newly identified miR-154-5p/KIF14 interaction offers further insights into the progression of NPC, which may represent a novel target for NPC diagnosis and treatment.
Keywords: MiR-154-5p, KIF14, nasopharyngeal carcinoma, migration and invasion
Introduction
Nasopharyngeal carcinoma (NPC) is the most common head and neck malignancy develops from the epithelial cells of the nasopharynx.1 NPC is characterized by unique genetic, highly metastatic feature and unique geographical distribution, with the highest incidence rate among the Cantonese living in Southern China, Southeast Asia.2 NPC has a high possibility of distant metastasis, while more than 70% of patients develop regional lymph node metastasis at the time of diagnosis, almost 20% of patients develop distant metastasis, mainly to the lungs or liver.3 It is universally accepted that NPC is highly sensitized to radiation therapy (RT), which is the main treatment for NPC.4 The major pattern of treatment failure is distant metastasis even after combined chemo-radiotherapy.5 Distant metastasis remains an arduous problem to be resolved in NPC.6,7 Hence, understanding the underlying molecular mechanisms of invasive and metastatic properties of NPC is of great importance.
MicroRNAs (miRNAs) are a class of short non-coding RNAs about 18–24 nt in size which are reported as key regulators of gene expression.8 MiRNAs have been found to play important roles in the development and progression of numerous tumors over the past decade, including NPC.9–11 They can act as oncogenic miRNAs or tumor suppressor miRNAs in the progression of cancer according to their downstream targets.12 Studies have shown that dysregulated miRNAs can affect tumor cell proliferation, migration, invasion and cancer metastasis by regulating related signal transduction pathways.13 Recent years, more and more miRNAs have been reported to function in NPC migration and invasion, such as miR-203a-3p,14 miR-23a15 and miR-3188.16
MiR-154-5p is a miRNA that has been reported to be dysregulated in many malignancies including NSCLC,17 skin squamous cell carcinoma18 and gastric cancer.19 It was revealed that MiR-154 was downregulated in gastric cancer and inhibited the invasion and growth of tumor cells by targeted DIXDC1 thus suppressed the activation of WNT signaling.19 Another study conducted by Zhao D revealed that miR-154 functions as a tumor suppressor in glioblastoma by targeting Wnt5a.20 A study conducted by Bruce et al21 has shown that miR-154-5p expression was associated with metastasis of NPC. However, the role and potential molecular mechanism of miR-154-5p in NPC migration and invasion is still undiscovered.
In the present study, we investigated the effect of miR-154-5p on migration and invasion in NPC. We found that miR-154-5p was downregulated in NPC tissues and negatively correlated with clinical N stages. MiR-154-5p could suppress cell invasion and migration. Additionally, the Kinesin-like protein KIF14 was identified as a new functional target of miR-154-5p. According to our results, we propose that miR-154-5p is a potential miRNA target for modulating the invasion and metastasis of NPC.
Materials and Methods
Patients and Samples
Forty cases of frozen NPC tissue and eight normal nasopharyngeal epithelial tissues were collected from the Radiotherapy Center of Jiangsu Cancer Hospital (Nanjing, China). All the patients included in the study provided written informed consent in accordance with the Declaration of Helsinki. All tissue samples were pathologically confirmed, and these clinical specimens were approved by the Ethics Review Committee of Jiangsu Cancer Hospital before being used in the study.
Cell Culture
Seven human NPC cell lines (5-8F, SUNE-1, 6-10B, CNE-1, CNE-2, HNE-1 and C666-1) and human immortalized nasopharyngeal epithelial cell lines (NP69) were provided by the Laboratory of Radiotherapy, Jiangsu Cancer Hospital. Among the cell lines, 5-8F, CNE-1, CNE-2, SUNE-1, 6-10B, HNE-1 and C666-1 were cultured in RPMI 1640 (Corning, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA); NP69 was cultured in serum-free medium (Invitrogen) containing bovine pituitary extract (BD Biosciences). All cell lines were grown in humid air at 37°C with 5% CO2. All the cell lines were approved by the Institutional Ethical Review Board of Jiangsu Cancer Hospital. Additionally, the cell lines we used were authenticated by STR profile.
RNA Extraction and qRT-PCR
Total RNA was extracted from NPC cells or clinical samples using TRizol reagent (Invitrogen). QRT-PCR reactions were performed on a 7500 FAST real-time PCR machine (Applied Biosystems) using SYBR Green PCR Master Mix. β-actin and U6 were used as standardized controls. The sequences of all primers are listed in Table S1.
Transfection
MiR-154-5p mimic, mimic negative control (mimic NC), miR-154-5p inhibitor, inhibitor negative control (inhibitor NC), KIF14 siRNA (si-KIF14) and siRNA negative control (si-NC) were synthesized by RiboBio (Guangzhou, China). Cell transfection was conducted using Lipofectamine 2000 (Invitrogen, CA, USA) according to the manufacturer’s instructions. After 48hrs of transfection, cells were harvested for qRT-PCR analysis and cell function experiments.
Colony Formation and Cell Viability Assay
Colony formation measurement: After transfection, spread in six-well plates at a density of 800 cells per well, discard the culture solution after 7–10 days of culture, fix with 4% paraformaldehyde for 30 mins, and stain with crystal violet 2 hrs. The colonies were observed and counted using Image J. Cell viability assay: CCK8 assay (Promega) was used to measure cell viability. After transfection, 3000 cells per well were seeded in 96-well plates. Then, CCK-8 reagent was added to the cultured cells. After 1 hr of incubation, each well was examined with a spectrophotometer at 450 nm.
Wound Healing Assay
Wound healing assay was performed to determine the ability of cells to migrate. The transfected cells were placed in a six-well plate, and cultured in a serum-free medium for 24 hrs, followed by scratching in parallel with a 200 μL pipette and washing free floating cells with PBS. Cell migration was measured at 0 and 24 hr under a light microscope at 100X magnification.
Transwell Assay
Transwell assays were performed according to the manufacturer’s protocol (BD Biosciences, Bedford, MA, USA) using Transwell chambers (for migration assays) or Matrigel pre-coated Transwell chambers (for invasion assays). Forty-eight hours after transfection, cells were collected, and these cells were resuspended (2×104 cells per well) into 200 μL of serum-free medium and plated in the upper chamber. Add 500 μL of RPMI 1640 with 20% FBS to the lower compartment. After 36 hrs of incubation, the upper chamber membrane was fixed and stained. An inverted microscope was used to count invading cells.
Luciferase Reporter Assay
The KIF14 wild-type (Wt) and mutant (Mut) 3ʹ-UTR were constructed and cloned into a pcDNA3.1 luciferase reporter vector. The NPC cells were cotransfected with KIF14 Wt or Mut 3ʹ-UTR vector and miR-154-5p mimic or mimic NC by using Lipofectamine 2000 reagent (Invitrogen). Two micrograms of a specific plasmid and mimic was cotransfected in NPC cells. After 48 hrs, renilla and firefly luciferase activities were assessed using the Dual-Luciferase Assay Kit (Promega) according to the Kit instructions.
Western Blot Analysis
Protein was extracted by lysing the treated cells using modified RIPA buffer and PMSF (Beyotime, Shanghai, China). A BCA protein assay kit was used to evaluate protein concentrations. Protein (10 mg) from each sample was separated using 10% SDS-PAGE gels and then transferred onto a nitrocellulose membrane. The membranes were incubated with specific antibodies, namely, anti-KIF14 (1:2000; Abcam HK, ab71155) and anti-β-actin (1:2000; Cell Signaling Technology, USA). Immunoreactive bands were imaged by using ECL detection reagents (Millipore, Billerica, MA, USA).
Tumor Xenograft Model
Six-week-old male BALB/c nude mice with immunodeficiency were obtained from GemPharmatech Co, Ltd (Jiangsu, China). To perform plantar tumor growth assays, green fluorescent protein (GFP) -labeled 5-8F cell suspensions were injected into the footpads of nude mice. When the tumor grew to about 100 mm3, the two groups were injected with the corresponding agent. The tumor volume was measured every 3–4 days and calculated using the following formula: V=ab2/2 (V, volume; a, longitudinal diameter; b, lateral diameter). 4–6 weeks after injection, all mice were sacrificed and dissected. The xenograft tumors were weighted. GFP was used to identify mice with lymph node metastasis. Animal experiments were conducted in accordance with the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals and followed protocols approved by the Animal Science Committee of Origin, Nanjing, China.
Immunohistochemistry
The expression of KIF14 in mouse tumour tissues was analysed on 2 μm thick, formalin-fixed and paraffin-embedded specimen sections. The slides with mounted tissue specimens were incubated in xylene for 5 mins, washed twice in 100% ethanol for 15 mins and then washed in 95% ethanol for 15 mins. Antigen unmasking was performed, and the slides were then blocked with 3% hydrogen peroxide for 20 mins at room temperature. Then, the primary antibody incubated at 4°C overnight, and then the kit (ZSGB BIO Inc) was used for the DAB chromogen followed by nuclear staining using haematoxylin.
Statistical Analysis
All statistical analyses were performed by Prism 7 (San Diego, CA, USA) and SPSS 23.0 (Chicago, IL, USA), including analysis of ANOVA, Student’s t-test and Chi-square test. Data are presented as mean±SD of three independent experiments, and P values <0.05 are considered statistically significant (* P <0.05, ** P <0.01 and *** P <0.001).
Results
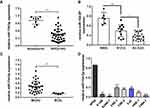
MiR-154-5p Is Down-Regulated in NPC Cell Lines and Clinical Specimens
Previous study has shown that miR-154-5p expression was associated with metastasis of nasopharyngeal carcinoma.21 To investigate whether miR-154-5p was abnormally expressed in nasopharyngeal carcinoma, we used qRT-PCR technology to detect miR-154-5p expression in 40 fresh frozen nasopharyngeal carcinoma tissues and 8 normal nasopharyngeal epithelial tissues. The results showed that the expression of miR-154-5p was significantly higher in nasopharyngeal carcinoma tissue than normal nasopharyngeal tissue (Figure 1A). Next, the correlation between miR-154-5p levels and some clinical features in 40 nasopharyngeal carcinoma tissues was analyzed. The analysis showed that the expression level of miR-154-5p was negatively correlated with clinical N stage. In the NPC tissues with high clinical N stage (N2-3), the level of miR-154-5p was significantly lower than that of low N stage (N0-N1) (Figure 1B). In addition, miR-154-5p expression in metastatic nasopharyngeal carcinoma tissues (M1) was lower than in non-metastatic (M0) (Figure 1C). Finally, we also found that miR-154-5p was downregulated in nasopharyngeal carcinoma cell lines compared to normal nasopharyngeal epithelial cells NP69 (Figure 1D). These data suggested that miR-154-5p was down-regulated in NPC and may function as a tumor suppressor.
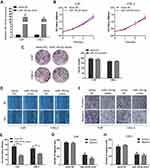
MiR-154-5p Inhibits NPC Cell Migration and Invasion in vitro
To evaluate the biological role of miR-154-5p in NPC, we transfected miR-154-5p mimic or mimic-NC into 5-8F and CNE-2 cells. The transfected efficiency revealed that the expression of miR-154-5p was upregulated in cells transfected with miR-154-5p mimic (Figure 2A). Colony formation, CCK8 assay, wound healing and transwell assay were used to detect cell proliferation and migration abilities. As shown in Figure 2B and C, the colony formation and proliferation ability of 5-8F and CNE-2 cells with miR-154-5p overexpression were insignificantly changed compared with the control groups. However, wound healing assay (Figure 2D and E), migration and invasion assay (Figure 2F–H) showed that miR-154-5p overexpression obviously suppressed NPC cell migration and invasion in vitro. These results indicated that miR-154-5p overexpression could inhibit the migration and invasion of NPC cells in vitro.
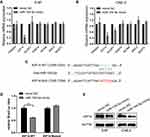
KIF14 Is a Direct Target of miR-154-5p in NPC Cells
In order to further explore the way in which miR-154-5p regulated the malignant progression of NPC, we performed bioinformatics analysis to predict downstream target genes of miR-154-5p and filtered 4478 and 169 genes, respectively, by using publicly available databases TargetScan and miRWalk. Finally, we used the 2 gene sets and selected the top seven candidate genes (TRIM, KIF14, NUSP1, RAB14, MCM6, SMC4, and MASTL) with higher comprehensive scores for further research. To verify the results, the mRNA level of seven target genes was detected after overexpression of miR-154-5p in 5-8F and CNE-2 cells, and only KIF14 was downregulated in both cells (Figure 3A and B). As predicted by bioinformatics analysis, KIF14 contained a binding site for miR-154-5p in its 3ʹ-UTR (Figure 3C). To validate the targeting of miR-154-5p to KIF14, a dual-luciferase reporter assay was performed. We constructed luciferase reporter vectors that included wild-type (WT) and mutated KIF14 3′-UTR. We then co-transfected these reporters with miR-154-5p mimic or mimic control in 5-8F cells. Results showed that overexpression of miR-154-5p inhibited the luciferase activity of the WT group compared with the mutation group (Figure 3D). To further determine whether miR-154-5p can regulate the KIF14 expression, we used Western blot to detect the expression of KIF14 in 5-8F and CNE-2 cells after transfection with miR-154-5p mimic. The results showed that miR-154-5p mimic inhibited the KIF14 expression (Figure 3E). All these results showed that KIF14 was a direct target of miR-154-5p and negatively regulated by miR-154-5p.
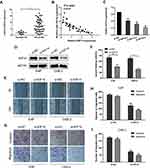
KIF14 Promotes NPC Cell Invasion and Migration in vitro
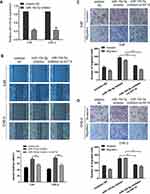
We subsequently explored the role of KIF14 in NPC cells. QRT-PCR was used to detect KIF14 mRNA expression in 40 nasopharyngeal carcinoma tissues and 8 normal nasopharyngeal tissues. The results showed that KIF14 expression at mRNA level was much higher in NPC tissues than normal tissues (Figure 4A). Moreover, KIF14 levels in NPC tissues were conversely related to miR-154-5p levels (Figure 4B). Next, we designed three siRNAs products and selected si-KIF14-3 which had the best knockdown efficiency for further functional study (Figure 4C). 5-8F and CNE-2 cells were treated with si-KIF14 or si-NC. KIF14 knockdown in the cells was detected through Western blot analysis (Figure 4D). Then, wound healing assays (Figure 4E and F) and transwell test (Figure 4G–I) were performed to verify the effect of KIF14 on the invasion and migration ability of NPC cells. The results showed that the invasion and migration capabilities of NPC cells were significantly inhibited when knocking down KIF14 expression.
Silencing KIF14 Reverses the Effects of miR-154-5p Expression Inhibition on NPC Cell Invasion and Migration
To further determine that KIF14 is the functional target of miR-154-5p in NPC, rescue experiments were performed. First, we designed and synthesized miR-154-5p inhibitor. 5-8F and CNE-2 cells were transfected with miR-154-5p inhibitor, which significantly decreased miR-154-5p expression (Figure 5A). Then, we co-transfected miR-154-5p inhibitor and si-KIF14 into the 5-8F and CNE-2 cells. Wound healing assays (Figure 5B) and transwell assays (Figure 5C and D) indicated that the miR-154-5p expression inhibition significantly increased cell migration and invasion ability, while silencing of KIF14 decreased the migration and invasion ability on the base of knockdown of miR-154-5p. Thus, miR-154-5p can affect tumor migration and invasion by regulating KIF14 expression in NPC cells.
MiR-154-5p Inhibits Tumor Metastasis in NPC in vivo
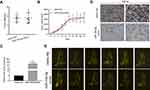
Based on the results in vitro, we further determined the effects of miR-154-5p on tumor metastasis in vivo. We constructed a model of spontaneous lymph node metastasis. MiR-154-5p overexpression vector or control vector was injected into the xenograft tumors twice a week. At the same time, we measured the tumor volume every 3–4 days. After 4–6 weeks of injection, the mice were sacrificed. We weighed the mass of the primary tumor and found no significant difference in tumor weight between the two groups (Figure 6A). Results also show that overexpression of miR-154-5p had no significant effect on the tumor volume compared with the control group (Figure 6B). In addition, the expression of miR-154-5p in tumour tissues was detected by qRT-PCR. The results showed that miR-154-5p expression was much higher in the miR-154-5p mimic group than it was in the control group (Figure 6C). Popliteal lymph node metastasis was observed after necropsy. In the mimic NC group, lymph node metastasis was observed in 5/6 rats at week 6 following implantation. In the miR-154-5p mimic group, lymph node metastasis was observed in 1/6 rats at week 6 following implantation. Compared with the respective controls, metastatic popliteal lymph nodes were significantly fewer in miR-154-5p mimic group (Figure 6E). The differences between the two groups were statistically significant (Table S2). Moreover, we used immunohistochemical staining to confirm the expression levels of KIF14 in popliteal metastatic nodes. Our results showed the decreased KIF14 expression in the miR-154-5p group compared with the control group (Figure 6D). Overall, these data suggested that miR-154-5p inhibits tumor metastasis in NPC in vivo.
Discussion
Great technological progress has been achieved in radiation therapy over the past decades, which greatly improve the outcome of NPC patient, with an 8-year overall survival (OS) of 68.5% and progression-free survival (PFS) of 62.6% in total patients as reported by Au et al.22 However, the N stage still regarded as a single adverse prognostic factor for NPC regional control, and distant metastasis is the most common form of treatment failure.23,24 Therefore, a better and clearer understanding of the molecular mechanism of NPC invasion and metastasis is needed.
In this study, we initially found significantly decreased miR-154-5p expression in NPC tissues compared to normal nasopharyngeal tissue. Additionally, we identified miR-154-5p was downregulated in NPC cell lines against NP69, the immortalized nasopharyngeal epithelial cell line. MiR-154-5p expression was negatively correlated with N and M state of NPC clinical feature, indicating that decreased levels of miR-154-5p may play an important role in invasion and metastasis of NPC. Interestingly, miR-154-5p significantly suppressed the migration and invasion ability of NPC cells in vitro. Overexpression of miR-154-5p by transfecting miR-154-5p mimic suppressed NPC metastasis in the established experimental xenograft model. However, little was known about the regulatory mechanisms for miR-154-5p in NPC migration, invasion and metastasis. Mechanistic studies demonstrated that miR-154-5p targets KIF14 directly by binding to the 3ʹ end regions (3ʹ-UTR) of KIF14 mRNA. Specifically, miR-154-5p mimic transfection reduced both mRNA and protein expression of KIF14 in both 5-8F and CNE-2 NPC cells.
KIF14, known as Kinesin-like protein 14, is a member of the kinesin protein family.25 As a microtubule motor protein, KIF14 can bind to microtubules and has a high ATPase activity.26,27 KIF14 is shown to play a key role in many cellular processes such as cytokinesis, cell division, proliferation, and apoptosis.28,29 In fact, it has been shown that KIF14 involved in the invasion and metastasis of several cancers.30 For example, Wang et al.31 reported that KIF14 promotes cancer cell proliferation through augmenting Akt phosphorylation in colorectal cancer. Another study conducted by Yang et al.32 revealed a similar result. Furthermore, they suggested a significant correlation between KIF14 expression and tumor node stage and metastasis stage, and KIF14 could be an independent prognostic factor for gastric cancer. In the current study, we identified KIF14 as a novel target of miR-154-5p in NPC from several candidates. MiR-154-5p binds to the 3ʹ-UTR of KIF14 mRNA and decreases the expression of KIF14 protein, which in turn suppresses NPC migration and metastasis.
In conclusion, our results provide evidence that miR-154-5p has a key role in regulating NPC cell migration, invasion and metastasis by targeting KIF14 and repressing its protein expression. Taken these together, our work provides new insights about the role of miRNAs in NPC progression and offers opportunities for application of miR-154-5p in therapeutic intervention as a novel target.
Acknowledgments
We thank the members of Jiangsu Cancer Hospital Radiotherapy Center for their technical assistance. This study was supported by the National Science Foundation of China (grant number 81872192; 81702685); Postdoctoral Science Foundation of China (grant number 2018M632266) and the Medical Young Talent Foundation of Jiangsu Provincial Health Department (grant number QNRC2016648).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Lee HM, Okuda KS, Gonzalez FE, Patel V. Current perspectives on nasopharyngeal carcinoma. Adv Exp Med Biol. 2019;1164:11–34. doi:10.1007/978-3-030-22254-3_2
2. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. doi:10.1016/S0140-6736(15)00055-0
3. Zhang B, Li MM, Chen WH, et al. Association of chemoradiotherapy regimens and survival among patients with nasopharyngeal carcinoma: a systematic review and meta-analysis. JAMA Net Open. 2019;2(10):e1913619. doi:10.1001/jamanetworkopen.2019.13619
4. Shuang H, Feng J, Caineng C, et al. The value of radical radiotherapy in the primary tumor of newly diagnosed oligo-metastatic nasopharyngeal carcinoma patients. Clin Transl Oncol. 2019;21(2):213–219. doi:10.1007/s12094-018-1911-7
5. Zeng L, Zhang Q, Ao F, et al. Risk factors and distribution features of level IB lymph nodes metastasis in nasopharyngeal carcinoma. Auris Nasus Larynx. 2019;46(3):457–464. doi:10.1016/j.anl.2018.10.012
6. Leong YH, Soon YY, Lee KM, Wong LC, Tham IWK, Ho FCH. Long-term outcomes after reirradiation in nasopharyngeal carcinoma with intensity-modulated radiotherapy: a meta-analysis. Head Neck. 2018;40(3):622–631. doi:10.1002/hed.24993
7. Almobarak AA, Jebreel AB, Abu-zaid A. Molecular targeted therapy in the management of recurrent and metastatic nasopharyngeal carcinoma: a comprehensive literature review. Cureus. 2019;11(3):e4210. doi:10.7759/cureus.4210
8. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi:10.1038/s41580-018-0059-1
9. Wu J, Hann SS. Functions and roles of long-non-coding RNAs in human nasopharyngeal carcinoma. Cell Physiol Biochem. 2018;45(3):1191–1204. doi:10.1159/000487451
10. Rupaimoole R, Calin GA, Lopez-Berestein G-B, Ak S. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. doi:10.1158/2159-8290.CD-15-0893
11. Guo H, Huang S, Li S, Yu H, Wu S, Zhou X. Prognostic significance of the long noncoding RNAs in nasopharyngeal carcinoma: a systematic review and meta-analysis. Cancer Manag Res. 2018;10:1763–1779. doi:10.2147/CMAR.S164695
12. Wang S, Claret FX, Wu W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma. Front Oncol. 2019;9:756. doi:10.3389/fonc.2019.00756.
13. Slack FJ, Chinnaiyan AM. The role of non-coding RNAs in oncology. Cell. 2019;179(5):1033–1055. doi:10.1016/j.cell.2019.10.017
14. Jiang N, Jiang X, Chen Z, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2017;36(1):138. doi:10.1186/s13046-017-0604-3
15. Bao L, You B, Shi S, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene. 2018;37(21):2873–2889. doi:10.1038/s41388-018-0183-6
16. Zhao M, Luo R, Liu Y, et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7(1):11309. doi:10.1038/ncomms11309
17. Lingling J, Xiangao J, Guiqing H, Jichan S, Feifei S, Haiyan Z. SNHG20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a miR-154 sponge. Biomed Pharmacother. 2019;112:108648. doi:10.1016/j.biopha.2019.108648
18. Chen H-Q, Gao D. Inhibitory effect of microRNA-154 targeting WHSC1 on cell proliferation of human skin squamous cell carcinoma through mediating the P53 signaling pathway. Int J Biochem Cell Biol. 2018;100:22–29. doi:10.1016/j.biocel.2018.04.021
19. Song J, Guan Z, Li M, et al. MicroRNA-154 inhibits the growth and invasion of gastric cancer cells by targeting DIXDC1/WNT signaling. Oncol Res. 2018;26(6):847–856. doi:10.3727/096504017X15016337254632
20. Zhao D, Wang R, Fang J, et al. miR-154 functions as a tumor suppressor in glioblastoma by targeting Wnt5a. Mol Neurobiol. 2017;54(4):2823–2830. doi:10.1007/s12035-016-9867-5
21. Bruce JP, Hui ABY, Shi W, et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget. 2015;6(6):4537–4550. doi:10.18632/oncotarget.3005
22. Au KH, Ngan RKC, Ng AWY, et al. Treatment outcomes of nasopharyngeal carcinoma in modern era after intensity modulated radiotherapy (IMRT) in Hong Kong: a report of 3328 patients (HKNPCSG 1301 study). Oral Oncol. 2018;77:16–21. doi:10.1016/j.oraloncology.2017.12.004
23. Lee AWM, Ng WT, Chan JYW, et al. Management of locally recurrent nasopharyngeal carcinoma. Cancer Treat Rev. 2019;79:101890. doi:10.1016/j.ctrv.2019.101890
24. Blanchard P, Nguyen F, Moya-plana A, et al. New developments in the management of nasopharyngeal carcinoma. Cancer Radiother. 2018;22(6–7):492–495. doi:10.1016/j.canrad.2018.06.003
25. Huang W, Wang J, Zhang D, et al. Inhibition of KIF14 suppresses tumor cell growth and promotes apoptosis in human glioblastoma. Cell Physiol Biochem. 2015;37(5):1659–1670. doi:10.1159/000438532
26. Arora K, Talje T, Asenjo A, et al. KIF14 binds tightly to microtubules and adopts a rigor-like conformation. J Mol Biol. 2014;426(17):2997–3015. doi:10.1016/j.jmb.2014.05.030
27. Rice S. Structure of kif14: an engaging molecular motor. J Mol Biol. 2014;426(17):2993–2996. doi:10.1016/j.jmb.2014.06.008
28. Singel S, Cornelius C, Zaganjor E, et al. KIF14 promotes AKT phosphorylation and contributes to chemoresistance in triple-negative breast cancer. Neoplasia. 2014;16(3):247–256, 256.e242. doi:10.1016/j.neo.2014.03.008
29. Zi B, Audusseau M, Riparbelli MG, Callaini G, D’avino PP. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc Natl Acad Sci U S A. 2013;110(24):9782–9787. doi:10.1073/pnas.1301328110
30. Singel S, C C, Batten K, et al. A targeted RNAi screen of the breast cancer genome identifies KIF14 and TLN1 as genes that modulate docetaxel chemosensitivity in triple-negative breast cancer. Clin Cancer Res. 2013;19(8):2061–2070. doi:10.1158/1078-0432.CCR-13-0082
31. Wang -Z-Z, Yang J, Jiang B-H, et al. KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int J Oncol. 2018;53(5):1939–1952. doi:10.3892/ijo.2018.4546
32. Yang Z, Li C, Yan C, et al. KIF14 promotes tumor progression and metastasis and is an independent predictor of poor prognosis in human gastric cancer. Biochimica et Biophysica Acta. Molecular Basis of Disease. 2019;1865(1):181–192. doi:10.1016/j.bbadis.2018.10.039
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.