Back to Journals » OncoTargets and Therapy » Volume 11
MiR-152 functioning as a tumor suppressor that interacts with DNMT1 in nasopharyngeal carcinoma
Authors Lu Z, Du M, Qian L, Zhang N, Gu J, Ding K, Wu J, Zhu H, He X , Yin L
Received 18 October 2017
Accepted for publication 17 January 2018
Published 27 March 2018 Volume 2018:11 Pages 1733—1741
DOI https://doi.org/10.2147/OTT.S154464
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Zhi-Wei Lu,1,2,* Ming-Yu Du,2,* Lu-Xi Qian,1,2 Nan Zhang,2 Jia-Jia Gu,2 Kai Ding,3 Jing Wu,2 Hong-Ming Zhu,2 Xia He,1,2 Li Yin1,2
1The Fourth Clinical School of Nanjing Medical University, Nanjing, Jiangsu, China; 2Department of Radiation Oncology, Jiangsu Cancer Hospital, Jiangsu Institute of Cancer Research, Nanjing Medical University Affiliated Cancer Hospital, Nanjing, Jiangsu, China; 3Department of Radiation Oncology, Suqian First Hospital, Suqian, Jiangsu, China
*These authors contributed equally to this work
Background: In recent years, miR-152 has been dysregulated in a variety of tumors and used as a tumor suppressor. Nevertheless, its role in nasopharyngeal carcinoma (NPC) remains unidentified.
Materials and methods: Real-time quantitative PCR (polymerase chain reaction) was performed to analyze the expression of miR-152 in NPC cell lines. MiR-152 expression profiles in NPC tissues were obtained from Gene Expression Omnibus (GEO GSE36682). The effect of miR-152 on the invasion and proliferation of NPC cells was determined through cell invasion, wound healing, and cell viability assays. Apoptosis was examined by flow cytometry, and Western blot was performed to measure expression of the target gene. Pyrosequencing was used to detect the methylation level of NPC cells.
Results: In this study, miR-152 was downregulated in the NPC tissues and cell lines. When miR-152 was enhanced, the invasion and migration of NPC cells were inhibited. However, miR-152 had no effect on the proliferation of NPC cells. Luciferase reporter gene analysis was performed, and the results showed that DNMT1 (DNA (cytosine-5)-methyltransferase 1) is a direct target of miR-152 in NPC cells. DNMT1 downregulation and miR-152 overexpression both reversed the effects of miR-152 inhibition on the NPC cells. In addition, miR-152 expression increased as a result of the inhibition of the methylation level of miR-152 when DNMT1 expression was downregulated.
Conclusion: The overexpression of miR-152 inhibited the migration and invasion of NPC cells by targeting DNMT1. Furthermore, DNMT1 regulated miR-152 expression through DNA methylation. Overall, the novel miR-152-DNMT1 regulatory circuit may provide better understanding of the pathogenesis of NPC and new epigenetic therapeutic target in NPC.
Keywords: miR-152, DNMT1, NPC, methylation
Introduction
Nasopharyngeal carcinoma (NPC) is a prevalent head and neck cancer in Southeast Asia, and is closely related to Epstein-Barr virus infection.1 Although NPC is sensitive to chemoradiation, the 5-year survival rates of individuals with NPC remain unsatisfactory. Local recurrence and distant metastasis are the main cause of failure.2,3 Therefore, the molecular mechanism of NPC pathogenesis must be explored.
MicroRNAs (miRNAs) are a group of highly conserved, noncoding, and single stranded small RNA (ribonucleic acid) molecules with lengths of 18–25 nucleotides. MiRNAs inhibit gene expression in combination with the 3′-UTR sequences of targeted mRNAs (messenger RNAs).4 Previous studies suggested that some miRNAs are involved in cancer formation, as they control cell functions, such as proliferation, differentiation, and apoptosis.5 So far, miR-152 is dysregulated in various tumors and tumor suppressors,6 such as gastric cancer,7 colorectal cancer,8 hepatocellular carcinoma,9 and prostate cancer.10 In addition, miR-152 is more downregulated in NPC than in normal tissue, indicating that it has potential as a tumor suppressor.11 Nevertheless, the role of miR-152 in NPC remains unknown.
DNMT1 (DNA (cytosine-5)-methyltransferase 1) is a type of methyltransferase responsible for maintaining DNA methylation during DNA replication.12 Several studies showed that DNMT1 expression considerably increases in NPC,13 and DNMT1 overexpression leads to an increase in DNA methylation; this increase is often associated with a poor prognosis of NPC.14 Furthermore, the upregulation of DNMT1 expression may promote DNA methylation, thereby reducing miR-152 expression.15 However, the relationship between miR-152 and DNMT1 in NPC remains unclear.
The purpose of this study is to investigate the role of miR-152 in the regulation, proliferation, invasion, and migration of NPC, and the mechanism of its interaction with DNMT1.
Materials and methods
Patients and samples
The MiR-152 expression profiles in NPC tissues were obtained from Gene Expression Omnibus (GEO GSE36682; www.ncbi.nlm.nih.gov/geo/). In addition, GEO2R was used for the comparison between the miR-152 expression levels of tumors and normal tissues.
Cell culture
Six NPC cells (CNE1, CNE2, 5-8F, 6-10B, HNE1, and SUNE1) and NP69 (ATCC, Manassas, VA, USA) were provided by the Research Center of Clinical Oncology of the Affiliated Jiangsu Cancer Hospital, Nanjing Medical University, in Nanjing, China. The cell lines were cultured in RPMI 1640 (Corning, Manassas, VA, USA) supplemented with 5% fetal bovine serum (FBS; Gibco, Waltham, MA, USA) at 37°C in a humidified chamber supplemented with 5% CO2.
RNA extraction and real-time qPCR
Total RNA was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Then, 2 μg of total RNA was reverse transcribed into cDNA with a reverse transcription reagent kit (Promega, Madison, WI, USA). The expression levels of mature miRNAs were amplified with SYBR Green quantitative real-time polymerase chain reaction (qRT-PCR) on an ABI7300 real-time PCR machine (Applied Biosystems, Waltham, MA, USA). The expression of miR-152 was normalized to the expression of U6. The primers for human miR-152 and U6 were synthesized by Ribobio (Guangzhou, China). The expression of DNMT1 was normalized to the expression of β-actin. The expression levels of DNMT1 and β-actin were examined, and the following specific primers were used: 5′-AGGCGGCTCAAAGATTTGGAA-3′ and 5′-GCAGAAATTCGTGCAAGAGATTC-3′ for DNMT1, and 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′ for β-actin. All reactions were performed in triplicate for each sample. Fold changes for miR-152-3p and DNMT1 expression levels were calculated through the 2-ΔΔCt method.
Cell transfection
MiR-152-3p mimic/inhibitor and miR negative control (NC) were synthesized by RiboBio. DNMT1-siRNA and NC-siRNA were obtained from RiboBio. CNE-2 cells were transfected with miRNA mimic/inhibitor/DNMT1-siRNA with Lipofectamine 2000 (Invitrogen) for 48 h. To investigate the relationship between miR-152 and DNMT1, MiR-152 inhibitor/NC inhibitor and DNMT1-siRNA/NC-siRNA were co-transfected into CNE-2 cells at the same time. To determine the transfection efficiency of miR-152 mimic/inhibitor/DNMT1-siRNA, the expression of miR-152/DNMT1 was assessed with a qRT-PCR detection system and Western blot apparatus.
Cell viability assay
Cell proliferation was assessed by cell counting kit-8 (CCK-8; Dojindo, Kumamoto Prefecture, Kyushu, Japan). After 48 h of transfection, cells were collected and reseeded in 96-well plates (3×103 cells per well) for cell viability assay. The reaction solution (10 μL) was added into cultured cells in 100 μL of culture medium and incubated at 37°C for 40 min. Cell proliferation was detected at 24, 48, and 72 h with CCK-8 (Dojindo). Optical density was determined at a wavelength of 450 nm.
Cell wound healing assay
We collected CNE-2 cells in logarithmic phase, and seeded them into six-well plates (1.5×105 cells per well). When cells reached 80%–90% confluence, similar-sized wounds were scratched on the surface of the well with 200 μL sterile micropipette tips. Cells were then washed three times with phosphate-buffered saline (PBS) and incubated in Dulbecco’s Modified Eagle’s Medium with 1% fetal bovine serum (FBS). Photographs were taken at indicated times (0 and 24 h) using an inverted microscope (Olympus, Tokyo, Japan).
Cell invasion assay
Invasion assays were performed with Transwell chambers, each with a pore size of 8 μm (BD Biosciences, San Jose, CA, USA). Transfected CNE-2 cells were cultured for 48 h. The cells in 200 μL of serum-free media were added to the upper compartment (2×104 cells per well) of the chamber (Corning), and the lower compartment was filled with 500 μL of RPMI-1640 and 20% FBS. After 36 h of incubation, the cells on the upper surface were removed, whereas the cells that migrated to the bottom surface were fixed in 4% methanol and stained with crystal violet. Six random fields of cells were counted in each well under a microscope at a magnification of 100×.
Flow cytometric analysis
After 48 h of transfection, CNE-2 cells were trypsinized and collected at 48 h. Cell pellets were rinsed once with PBS and fixed overnight in 70% ice-cold ethanol at −20°C. The cells were then resuspended in PBS, stained with propidium iodide containing RNase A for 30 min at 37°C, and analyzed by flow cytometry. Each assay was repeated at least three times.
Western blot analysis
After 48 h of transfection, cells were extracted and prepared in modified radioimmunoprecipitation assay buffer (Beyotime, Shanghai, China). Total protein was extracted, and protein concentration was quantified with a bicinchoninic acid (BCA) protein assay kit (Beyotime). A total of 20 mg of protein from each sample was used for Western blot analysis. The primary antibodies used in this study included monoclonal anti-DNMT1 (1:1,000; Cell Signaling Technology, Danvers, MA, USA), and β-actin was used as loading control. Immunoreactive bands were visualized with ECL (enhanced chemiluminescence) detection reagent (Millipore, Billerica, MA, USA). All data analyses were repeated three times independently.
Luciferase reporter construction and luciferase assays
Human DNMT1 3′UTR regions containing predicted binding site for miR-152-3p were constructed on pmirGLO vectors, and a sequence of pri-miR-152-3p was inserted into pCDNA3.1 plasmid. The CNE-2 cells were inoculated into 24-well plates and cultured for 24 h. The cells were subsequently co-transfected with Wt/Mut reporter plasmid (0.2 μg) and miR-152 mimic/miR-NC. Cells were harvested 48 h after transfection. Luciferase activities were analyzed using the dual-luciferase reporter assay system according to manufacturer’s protocol.
Experiment with 5-aza-2′-deoxcycytidine
DNA-hypomethylating drug 5-aza-2′-deoxycytidine (Sigma-Aldrich, St Louis, MO, USA) was used to verify the regulation of DNA methylation in the NPC cells. The cells were treated with 5-aza-cdr (0, 10, and 20 μmol/L) for 3 days and with TSA (100 ng/mL) for an additional 24 h. Cells were harvested for DNA and RNA extraction.
DNA isolation and bisulfite conversion
Genomic DNA was isolated from CNE-2 cells using a QIAamp DNA Mini Kit (Qiagen, Venlo, the Netherlands) according to the manufacturer’s instructions. A Qiagen’s EpiTect bisulfite kit was used for the bisulfite conversion of the DNA sample, and 1–2 μg of DNA was diluted to 40 μL with deuterium depleted water in 0.5 mL PCR tubes. Freshly prepared bisulfite conversion (85 μL) and DNA protection solution (PB 15 μL) was then added to the solution. After mixing, the reaction tube was placed in PCR instrument at 95°C for 5 min and at 60°C for 1 h for five cycles. The bisulfite-converted DNA was stored at 20°C.
Pyrosequencing
The miR-152 primers for the pyrosequencing assays were designed in PyroMark® Assay Design SW 2.0. The design contained a sequencing primer and an amplification primer set in which one primer was biotinylated. Bisulfite-converted DNA was amplified with PyroMark® PCR kit on a thermal cycler as follows: 3 min at 95°C, 50 cycles of 15 s at 95°C, 20 s at 54°C, and 30 s at 72°C, then a 5 min final extension stage at 72°C. Subsequently, we isolated single-strand DNAs and performed pyrosequencing assays on PyroMark® Q96 according to the manufacturer’s instructions. The degree of methylation of the target gene locus was analyzed with PyroMark® Q96 Software (Sequencing section containing eight CpG sites: GGAGGCCACGCGCGAGAGGGGGCCGCGGGGCGGCGCGGAGG).
Statistical analysis
Student’s t-test and one-way ANOVA were performed in GraphPad Prism 5.0 software and SPSS 13.0. Data are expressed as mean and standard deviation from at least three separate experiments. Differences were considered significant at a P-value of <0.05.
Results
MiR-152 is downregulated in NPC tissues and cell lines
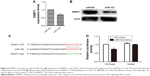
To confirm the role of miR-152 in tumorigenesis of NPC, we searched the GEO to obtain the expression profiles of miR-152 in NPC and normal tissues. As expected, the expression of miR-152 in NPC (n=62) tissues was significantly lower than in normal tissue (n=6), suggesting that the tumor suppressed miR-152 expression in NPC (GSE36682; Figure 1A). For the verification of the hypothesis, the mRNA levels of miR-152 in the six NPC cell lines (5–8F, 6–10B, HNE1, SUNE1, CNE-1, and CNE-2) and NP69 were determined by qRT-PCR (Figure 1B). MiR-152 expression decreased in the six NPC cells, similar to that in NPC tissues. In addition, CNE-2 had the lowest miR-152 expression in the NPC cell lines. Hence, we used CNE-2 cells as a model for subsequent experiments.
Overexpression of miR-152 inhibits invasion and migration of NPC cells
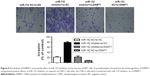
To clarify the role of miR-152 in NPC cells, we transfected CNE-2 cells with miR-152 mimics. By qRT-PCR, miR-152 expression considerably increased (Figure 2A). In the matrigel invasion assay, the number of cells invaded significantly decreased in the miR-152 mimic cells (Figure 2B). Furthermore, miR-152 had similar effects in the wound healing assays (Figure 2C). These results suggested that miR-152 inhibits the migration and invasion of NPC cells. However, cell proliferation was unaffected by the enhancement of miR-152 expression (Figure 2D). Similarly, no significant difference was observed between the mimic and control groups in the apoptosis assays (Figure 2E). In addition, when the expression of miR-152 was downregulated (Figure 2F), the invasion (Figure 2G) and migration (Figure 2H) of NPC cells was promoted.
Identification of DNMT1 as a direct target of miR-152 in NPC cells
Identifying a target gene is crucial to the exploration of the mechanism of miR-152 in NPC cells. DNMT1 was selected as a potential target gene for miR-152. TargetScan software was used for the screening. To verify the relationship between miR-152 and DNMT1, we transfected CNE-2 cells with miR-152 mimic or negative control. By increasing expression of miR-152, we observed the downregulation of DNMT1 expression through qRT-PCR (Figure 3A) and Western blot analysis (Figure 3B). In addition, the 3′UTR of DNMT1 contained a conserved binding site for miR-152 (Figure 3C). To confirm that DNMT1 is a target gene of miR-152-3p in NPC cells, we constructed three vectors (pmirGLO, DNMT1-3UTR-WT, and DNMT1-3UTR-MUT) for the implementation of subsequent luciferase assay. Co-transfection of miR-152 and DNMT1-3′-UTR-WT significantly reduced the activity of luciferase, although this effect was not achieved in the mutant group (Figure 3D). Based on the above results, DNMT1 is an miR-152 target gene.
DNMT1 is involved in invasion and migration of NPC cells
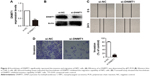
Although DNMT1 is a target for miR-152, and DNMT1 overexpression (31 of 32, 97%) can be observed in NPC tissues,13 the role of DNMT1 in NPC cells remains unclear. We transfected CNE-2 cells with DNMT1-siRNA or NC-siRNA, and performed qRT-PCR (Figure 4A) and Western blot experiments (Figure 4B) to verify whether DNMT1 expression was significantly reduced. After confirming the downregulation of DNMT1 expression, a matrigel invasion test and wound healing assays were conducted to validate the effect of DNMT1 on the invasion and migration ability of NPC cells. The results showed that the downregulation of DNMT1 expression significantly inhibited the migration (Figure 4C) and invasion (Figure 4D) of NPC cells.
Downregulation of DNMT1 reversing the effect of inhibition of miR-152 expression on invasion and migration of NPC cells
To further clarify that miR-152 regulates invasion of NPC cells, DNMT1, we co-transfected miR-152 inhibitor and DNMT1-siRNA into the CNE-2 cells. Then, miR-152 Inhibitor+si-NC, DNMT1-si+miR-152NC, and miR-152NC+si-NC were designed for comparison. The results of matrigel invasion experiments showed that the inhibition of miR-152 expression increased cell migration ability, and the simultaneous silencing of DNMT1 can reverse this effect (Figure 5). Thus, miR-152 can act as a tumor suppressor in NPC cells by modulating DNMT1.
DNMT1 regulates miR-152 expression through DNA methylation
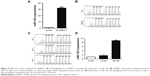
The regulatory circuit of miR-152 and DNMT1 was reported in several cancers, such as breast16 and prostate cancer.10 Silencing DNMT1 increased miR-152 expression in the CNE-2 cells (Figure 6A). Given that DNMT1 regulates methylation, we hypothesized that downregulating DNMT1 expression can attenuate the methylation level of miR-152 and consequently increases miR-152 expression. To verify this hypothesis, we transfected CNE-2 cells with DNMT1-SI. The methylation level of miR-152 was significantly decreased through pyrosequencing, and PCR results showed that miR-152 expression was elevated (Figure 6B). Furthermore, CNE-2 cells were treated with 5-aza-2′-deoxcycytidine. The DNA methylation level of miR-152 was gradually decreased with the increase of 5-aza-2′-deoxcycytidine concentration (0, 10, 20 μmol/L; Figure 6C), and its expression was upregulated as expected (Figure 6D). Consequently, our results show that the downregulation of DNMT1 expression inhibited the methylation level of miR-152 on its CpG islands, thereby increasing miR-152 expression.
Discussion
Owing to the developments in radiotherapy technology and treatment mode, local control and overall survival of NPC patients were effectively improved.17 However, the prognosis of 10%–20% patients with NPC remains poor because of local recurrence and distant metastasis.18,19 Exploring the molecular mechanisms of the invasion and metastasis in NPC may provide new effective strategies for the prevention, screening, and treatment of NPC.
Specific miRNAs are dysregulated in NPC, and play an important role in the progression and development of the disease.20 MiRNAs can be divided into two groups (according to their role in cancer), namely, oncogenes and tumor suppressor genes.5 Low miR-152 expression is frequently observed in most tumors; this condition suggests that miR-152 is a tumor suppressor in human cancer.6 However, miR-152 is upregulated in neuroblastoma and may act as an oncogene.21 In the present study, miR-152 expression was considerably reduced in NPC, as indicated by the GEO2R results. MiR-152 expression was downregulated in the six NPC cells. The upregulated miR-152 expression levels in the CNE-2 cells considerably inhibited cell invasion and migration, but had no significant effect on proliferation. We obtained opposite results when miR-152 expression was downregulated. All these results indicate that miR-152 plays a role as a tumor suppressor in NPC.
MiRNA exerts its function mainly by binding to the 3′UTR region of the target gene.22 In our study, we screened several target genes for miR-152 using TargetScan and finally selected DNMT1 according to the PCR results. DNMT1 is a methyltransferase and highly expressed in NPC.13 Previous studies showed that DNMT1 inhibits E-cadherin expression by increasing the rate of methylation of its promoter region, subsequently blocking NPC metastasis.23 In our study, increased miR-152 expression led to the downregulation of DNMT1 expression at mRNA and protein levels. We also confirmed that the knockdown of DNMT1 expression inhibited the invasion and migration of the NPC cells. This result was also observed when miR-152 expression was enhanced. Moreover, silencing of DNMT1 reverses the effect of miR-152 inhibition on the NPC cells. Luciferase assay results further confirmed that DNMT1 is the direct target of miR-152.
Abnormalities in genomic methylation may lead to the aberrant silencing of many tumor suppressor genes (TSGs), cellular functional genes, and miRNAs, and is, thus, closely related to the occurrence and development of cancer.14,24 Many publications revealed that miR-152, as a tumor suppressor, can be deactivated by methylation due to DNMT1.10,15 In the present study, the knockdown of DNMT1 increased miR-152 expression, indicating a negative feedback loop between miR-152 and DNMT1. Meanwhile, the level of miR-152 methylation decreased. Moreover, in the CNE-2 cells treated with 5-aza-2′-deoxcycytidine, the results were similar to those observed after knocking out DNMT1, but were more pronounced.
Increasing evidence showed that aberrant DNA methylation at promoter CpG islands of TSGs is common in NPC, particularly PCDH8,25 RASSF1A,26 and PCDH10.27 MiR-152 may increase TSG expression by targeting DNMT1. MiR-152 might increase TSG expression by targeting DNMT1 so that it is involved in the tumorigenesis of NPC. E-cadherin was one of those TSGs which could also be modulated by DNMT1 because of methylation at its promoter region.13 To our knowledge, the loss of membranous E-cadherin expression can result in enhanced cell migration activity, because E-cadherin acts as an adhesion molecule that regulates cell–cell contact.23,28 Moreover, this phenomenon is also closely related to the invasion and metastasis of NPC.29 In our study, miR-152 could inhibit the migration and invasion of NPC by targeting DNMT1. From our observations, we proposed a hypothesis that downregulation of DNMT1 caused by miR-152 might lead to enhanced expression of E-cadherin, which is responsible for the inhibition of migration and invasion in NPC.
Conclusion
MiR-152 is downregulated in NPC and functions as a tumor suppressor by targeting DNMT1. In addition, a novel miR-152-DNMT1 regulatory circuit may provide a new epigenetic therapeutic target in NPC.
Acknowledgments
We thank the members of the Research Center of Clinical Oncology of Jiangsu province for their technical assistance. This work was supported by the Jiangsu Provincial Commission of Health and Family Planning Young Scholars Award (grant numbers Q201501); the Jiangsu Clinical Medicine Science and Technology Special Fund (grant number BL2014091); the National Natural Science Foundation of China (grant number 81672989); and the Medical Young Talent Foundation of Jiangsu Provincial Health Department (grant number QNRC2016648).
Disclosure
The authors report no conflicts of interest in this work.
References
Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma – review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30(7):946–963. | ||
Zhang L, Chen QY, Liu H, Tang LQ, Mai HQ. Emerging treatment options for nasopharyngeal carcinoma. Drug Des Devel Ther. 2013;7:37–52. | ||
Bensouda Y, Kaikani W, Ahbeddou N, et al. Treatment for metastatic nasopharyngeal carcinoma. Eur Ann Otorhinolaryngol Head Neck Dis. 2011;128(2):79–85. | ||
Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. | ||
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. | ||
Liu X, Li J, Qin F, Dai S. miR-152 as a tumor suppressor microRNA: Target recognition and regulation in cancer. Oncol Lett. 2016;11(6):3911–3916. | ||
Zhai R, Kan X, Wang B, et al. miR-152 suppresses gastric cancer cell proliferation and motility by targeting CD151. Tumour Biol. 2014;35(11):11367–11373. | ||
Li B, Xie Z, Li B. miR-152 functions as a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumour Biol. 2016;37(8):10075–10084. | ||
Dang YW, Zeng J, He RQ, Rong MH, Luo DZ, Chen G. Effects of miR-152 on cell growth inhibition, motility suppression and apoptosis induction in hepatocellular carcinoma cells. Asian Pac J Cancer Prev. 2014;15(12):4969–4976. | ||
Theodore SC, Davis M, Zhao F, et al. MicroRNA profiling of novel African American and Caucasian Prostate Cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget. 2014;5(11):3512–3525. | ||
Li T, Chen JX, Fu XP, et al. microRNA expression profiling of nasopharyngeal carcinoma. Oncology Rep. 2011;25(5):1353–1363. | ||
Hermann A, Goyal R, Jeltsch A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J Biol Chem. 2004;279(46):48350–48359. | ||
Tsai CL, Li HP, Lu YJ, et al. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66(24):11668–11676. | ||
Jiang W, Cai R, Chen QQ. DNA methylation biomarkers for nasopharyngeal carcinoma: diagnostic and prognostic tools. Asian Pac J Cancer Prev. 2015;16(18):8059–8065. | ||
Xu Q, Jiang Y, Yin Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5(1):3–13. | ||
Sengupta D, Deb M, Rath SK, et al. DNA methylation and not H3K4 trimethylation dictates the expression status of miR-152 gene which inhibits migration of breast cancer cells via DNMT1/CDH1 loop. Exp Cell Res. 2016;346(2):176–187. | ||
Tang LL, Sun Y, Mao YP, et al. Prognostic value of parapharyngeal extension in nasopharyngeal carcinoma treated with intensity modulated radiotherapy. Radiother Oncol. 2014;110(3):404–408. | ||
Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. | ||
Lee N, Harris J, Garden AS, et al. Intensity-modulated radiation therapy with or without chemotherapy for nasopharyngeal carcinoma: radiation therapy oncology group phase II trial 0225. J Clin Oncol. 2009;27(22):3684–3690. | ||
Bruce JP, Liu FF. MicroRNAs in nasopharyngeal carcinoma. Chin J Cancer. 2014;33(11):539–544. | ||
Liu DZ, Ander BP, Tian Y, et al. Integrated analysis of mRNA and microRNA expression in mature neurons, neural progenitor cells and neuroblastoma cells. Gene. 2012;495(2):120–127. | ||
Huang T, Yin L, Wu J, et al. MicroRNA-19b-3p regulates nasopharyngeal carcinoma radiosensitivity by targeting TNFAIP3/NF-kappaB axis. J Exp Clin Cancer Res. 2016;35(1):188. | ||
Zhang RL, Peng LX, Yang JP, et al. IL-8 suppresses E-cadherin expression in nasopharyngeal carcinoma cells by enhancing E-cadherin promoter DNA methylation. Int J Oncol. 2016;48(1):207–214. | ||
Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. | ||
He D, Zeng Q, Ren G, et al. Protocadherin8 is a functional tumor suppressor frequently inactivated by promoter methylation in nasopharyngeal carcinoma. Eur J Cancer Prev. 2012;21(6):569–575. | ||
Ye M, Huang T, Ni C, Yang P, Chen S. Diagnostic capacity of RASSF1A promoter methylation as a biomarker in tissue, brushing, and blood samples of nasopharyngeal carcinoma. EBioMedicine. 2017;18:32–40. | ||
Ying J, Li H, Seng TJ, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25(7):1070–1080. | ||
Birchmeier W, Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim Biophys Acta. 1994;1198(1):11–26. | ||
Ran Y, Wu S, You Y. Demethylation of E-cadherin gene in nasopharyngeal carcinoma could serve as a potential therapeutic strategy. J Biochem. 2011;149(1):49–54. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.