Back to Journals » OncoTargets and Therapy » Volume 13
miR-145-5p Regulates the Proliferation, Migration and Invasion in Cervical Carcinoma by Targeting KLF5
Authors Cao H, Pan G, Tang S, Zhong N, Liu H, Zhou H, Peng Q, Zou Y
Received 6 December 2019
Accepted for publication 25 February 2020
Published 20 March 2020 Volume 2020:13 Pages 2369—2376
DOI https://doi.org/10.2147/OTT.S241366
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Hui Cao, Guihua Pan, Shiqiang Tang, Ni Zhong, Huake Liu, Haizhi Zhou, Qin Peng, Yongbin Zou
Department of Oncology, Chenzhou First People’s Hospital, Chenzhou, People’s Republic of China
Correspondence: Yongbin Zou
Department of Oncology, Chenzhou First People’s Hospital, 102 Luojiajing Road, Chenzhou 423000, People’s Republic of China
Tel +86-13507357355 Email [email protected]
Objective: Cervical carcinoma (CC) is a serious threat to women’s health and few effective therapeutic methods have been discovered. The purpose of this study is to explore the underlying mechanism of miR-145-5p in CC.
Methods: Bioinformatics methods were employed to analyze the gene expression data of CC from TCGA database. qRT-PCR was used to detect the expression of miR-145-5p and KLF5 in CC cells, and Western blot was employed for the examination of KLF5 protein level. The targeted relationship between miR-145-5p and KLF5 was verified by a dual-luciferase reporter assay. Moreover, CCK-8, wound healing assay and transwell invasion assay were used to analyze the effects of miR-145-5p overexpression or KLF5 silencing on the proliferation, migration and invasion of CC cells.
Results: miR-145-5p was shown to be down-regulated in CC tissues and cells, while KLF5 was up-regulated. miR-145-5p could bind to the complementary sequence within the wild type KLF5 3ʹUTR rather than the mutant one. In addition, miR-145-5p could effectively down-regulate KLF5, in turn inhibiting the proliferation, migration and invasion of CC cells.
Conclusion: miR-145-5p regulates the proliferation, migration and invasion of CC cells by targeting KLF5.
Keywords: cervical carcinoma, miR-145-5p, KLF5, proliferation, migration, invasion
Introduction
Cervical carcinoma (CC) is the second common cancer among women with higher morbidity and mortality.1,2 Almost 95% CC is caused by persistent infection of human papillomavirus (HPV).3,4 Despite the pivotal role in the transformation of cervical epithelial cells, HPV infection is not enough to result in the malignant conversion and consequently lead to tumor occurrence.5 In the infected bodies, CC progression can be affected by various auxiliary factors, especially by some differentially expressed genes.6,7 Therefore, knowing more about the abnormally expressed genes can help to find novel therapeutic methods for CC and improve patient’s prognosis.
MicroRNAs (miRNAs) have been found to be abnormally expressed in a variety of cancer types, and they can catalyze mRNA cleavage or inhibit translation in the way of interacting with the complementary sequences of target mRNAs, thus affecting cancer progression.8–10 Wu et al discovered that miR-144-3p was significantly down-regulated in CC, and it inhibited cell proliferation, migration and invasion by targeting MAKP611. Li et al found that miR-93-5p played an inhibitory role in the proliferation and metastasis of CC cells, and it could suppress the development of HPV-positive CC by targeting BTG3.12 These studies suggest that the abnormal expression of miRNAs may affect the malignant progression of CC. It has been reported that miR-145-5p is down-regulated in a variety of human cancers, such as breast cancer13 and bladder cancer14, and plays an inhibitory role in tumor growth. In addition, miR-145 has been confirmed to have the potential serving as a candidate biomarker for human diagnosis.15 Sathyanarayanan et al also found that miR-145 inhibited the proliferation and metastasis of CC cells by targeting SIP1.16 Moreover, some bioinformatics studies have reported that the down-regulation of miR-145 is associated with poor prognosis of CC.17,18 Therefore, it is meaningful to study the potential mechanism of miR-145-5p in CC. Generally, miRNAs function in cancer via their target genes. KLF5, a member of Krüppel-like transcription factor (KLF) family, participates in a variety of biological processes, such as cell proliferation, differentiation, growth, etc.19,20 Studies have shown that high KLF5 can promote cell growth and cause phenotypic changes, and it has been considered to be a novel target for cancer treatment.21,22 These studies collectively suggest that miR-145-5p and KLF5 may affect the malignant progression of CC.
Currently, about 70% of CC cases have been prevented by HPV vaccines (HPV-16 and HPV-18).19 In this study, we used the bioinformatics technique and in vitro experiments to clarify the molecular mechanism of the miR-145-5p/KLF5 axis underlying the occurrence and development of CC, and aimed to provide a theoretical support for the targeted therapy of CC.
Materials and Methods
Bioinformatics Analysis
Expression data of miRNAs and mRNAs of CC were accessed from TCGA database (https://cancergenome.nih.gov/). “edgeR” package was used to perform differential analysis (| logFC | > 2, Padj < 0.01) to obtain the differentially expressed miRNAs (DEmiRNAs) and mRNAs (DEmRNAs). Then survival analysis was conducted and confirmed miR-145-5p to be our research object. TargetScan, miRDB and miRTarBase three databases were applied to predict the downstream target mRNAs of miR-145-5p, and a Venn diagram was made to find the potential target mRNA.
Cell Culture and Transfection
Human CC cell lines C33A (HTB-31), HT-3 (HTB-32), Hela (CCL-2) and normal cervical cell line HcerEpic (AC340374) were cultured in the Dulbecco’s Modified Eagle Medium (DMEM; 30-2002) containing 10% fetal bovine serum (FBS), and maintained in a 37°C incubator containing 5% CO2. These cells and culture mediums were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA).
CC cells in logarithmic phase were divided into negative control (NC) mimic, miR-145-5p mimic, siRNA (si)-NC, si-KLF5, NC mimic + si-NC, miR-145-5p mimic + si-NC and miR-145-5p mimic + si-KLF5 groups. NC mimic, miR-145-5p mimic, si-NC and si-KLF5 (Ribo Bio; Guangzhou, China) were transiently transfected into cells using Lipofectamine ®2000 reagent (Thermo Fisher Scientific, Inc., Waltham, USA), respectively, and then the cells were cultured in corresponding mediums with 5% CO2 at 37°C. After 6 h, the culture mediums were changed and the cells were continuously cultured for further 48 h for follow-up experiments.
Real-Time Quantitative PCR (qRT-PCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen, Thermo Fisher Science, Inc.), and then reversely transcribed into cDNA by the SuperScript® II Reverse Transcriptase kit (Invitrogen, USA). qRT-PCR was performed using the SYBR® Premix Ex Taq™ (Takara, Japan) under the following thermal cycling conditions: pre-denaturation at 95°C for 10 min, 40 cycles at 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. U6 and GAPDH were applied as endogenous regulators. The relative expression levels of the target miRNA and mRNA were calculated using the 2−ΔΔCt method. The primers used were synthesized by the GeneCopoeia (Maryland, USA) and the sequences were as follows: miR-145-5p (F: 5ʹ-CTCACGGTCCAGTTCCCA-3ʹ; R: 5ʹ-ACCTCAAGAACAGTATTTCCAGG-3ʹ); U6 (F: 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ; R:5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ); KLF5 (F: 5ʹ-ACACCAGACCGCAGCTCCA-3ʹ; R: 5ʹ-TCCATTGCTGCTGTCTGATTTGTAG-3ʹ); GAPDH (F: 5ʹ-ACAGTCAGCCGCATCTTCTT-3ʹ; R: 5ʹ-ACGACCAAATCCGTTGACTC-3ʹ).The experiment was repeated 3 times independently.
Western Blot
Total proteins were isolated from cells with RIPA lysate buffer (P0013C, Beyotime, Jiangsu, China). BCA Protein Assay Kit (23227, Thermo Fisher Scientific) was used to determine the concentrations of the protein samples, and the samples were quantified according to different concentrations. Thereafter, the proteins were boiled for 10 min at 95 °C with 10 μL of loading buffer (1610737, Haoran Biological Technology, Shanghai, China) and then separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) at 100V. After electrophoresis, the proteins were transferred onto nitrocellulose membranes, which were sequentially blocked by 5% BSA/TBST for 60 min. Primary rabbit polyclonal antibodies KLF5 (ab137676, 1:2000, Abcam, Cambridge, USA) and GAPDH (ab181602, 1:1000, Abcam) were added onto the membranes for incubation overnight at 4°C, followed by secondary antibody goat anti-rabbit IgG H&L (ab216773, 1:1500, Abcam) at room temperature for 1 h. The membranes were washed with TBST buffer for 3 times. An Enhanced Chemiluminescence (ECL) kit (ECL808-25, Biomiga, USA) was used for the visualization of protein bands, and the Image Pro Plus 6.0 (Media Cybernetics, USA) software was applied to analyze the relative protein levels. The experiment was repeated 3 times independently.
Dual-Luciferase Reporter Gene Assay
The amplified sequences of the wild type 3ʹ UTR of KLF5 (WT-3ʹUTR) were cloned into the pmirGLO vectors (Promega Corp., WI, USA) for the construction of luciferase Wt-KLF5 vectors, and the mutant ones (luciferase Mut-KLF5) were synthesized by GenScript (Nanjing, China). miR-145-5p mimic and NC mimic were co-transfected with Wt-KLF5 or Mut-KLF5 into cells, respectively. After 48 h, Firefly and Renilla luciferase activities were measured using the Dual-Luciferase Reporter system (Promega) following the manufacturer’s instructions. The fluorescence intensity was measured by a luminometer TD-20/20 (E5311, Promega). The Firefly luciferase activity was normalized to Renilla luciferase activity for each sample. The experiment was repeated 3 times independently.
CCK-8
Cell Counting Kit-8 (CCK-8, Beyotime, Shanghai, China) was used to assess CC cell proliferation. Cell suspension were seeded into 96-well plates at a density of 8× 103 cells/well, and maintained in 5% CO2 at 37 °C. At 0, 24, 48, 72 and 96 h, 10 μL of CCK-8 reagent was added into each well for 4 h of incubation, respectively. Then, the absorbance values were measured at 450nm. The experiment was repeated in triplicate.
Transwell Invasion Assay
24-well Transwell inserts (BD, Biosciences, MD, USA) with 8μm in aperture were used to detect cell invasion. About 2 × 104 cells were added into the upper chambers that were pre-coated with Matrigel matrix (BD Bioscience), and RPMI-1640 medium containing 10% FBS was filled into the lower chambers as an attractant. After incubation at 37°C for 24 h, the cells that did not invade to the lower chambers were removed with cotton swabs, and the invaded cells were fixed and stained with 100% methanol and 0.1% crystal violet. Five fields of the view were randomly selected for cell count under a microscope. The experiment was repeated 3 times independently.
Wound Healing Assay
Wound healing assay was performed for the detection of cell migration. Cells were inoculated in 6-well plates and a scrape across the single layer was made with the tip of a 200μL pipette when the cell confluence reached 80%. The wells were briefly washed twice to remove the separated cells. Then, the cells were grown with fresh mediums. The scratch area was photographed with an inverted microscope after 0 h and 24 h, and the wound healing rate was calculated using the Image J. The experiment was repeated 3 times independently.
Statistical Analysis
The SPSS v.24.0 software (IBM Corp., NY, USA) was applied for statistical analysis, and the GraphPad Prism 6 software (GraphPad Software, Inc., CA, USA) was used for the completion of graphical demonstrations. All data were expressed as mean ± standard deviation (SD). Student’s t test was used for analyzing the comparisons between two groups, and one-way analysis of variance (ANOVA) was applied for the comparisons among multiple groups. P<0.05 was considered statistically significant.
Results
miR-145-5p Is Down-Regulated in CC Tissues and Cells
“edgeR” package was used to identify the DEmiRNAs and DEmRNAs in the TCGA-CESC dataset. As shown in Figure 1A, it was found that miR-145-5p was significantly down-regulated in cancer tissues. Survival analysis showed that low miR-145-5p was markedly correlated with prognosis of CC patients (Figure 1B). These results suggested that the down-regulation of miR-145-5p could lead to the tumor progression. Afterwards, miR-145-5p was further detected in human normal cervical cell line HcerEpic and human CC cell lines C33A, HT-3 and Hela by qRT-PCR (Figure 1C). It was found that miR-145-5p was remarkably decreased in cancer cases relative to the normal cell line, and the lowest expression was shown in Hela cells. Therefore, Hela cells were chosen for follow-up experiments.
KLF5 Is Up-Regulated in CC Tissues and Cells
In order to further understand the downstream regulatory mechanism of miR-145-5p in CC, we used miRDB, miRTarBase and TargetScan databases to predict the target mRNAs of miR-145-5p. The Venn diagram (Figure 2A) was made to find the overlapping mRNAs, among which KLF5 was with the putative binding sites with miR-145-5p. Subsequently, KLF5 level was determined in the TCGA-CESC dataset, finding that KLF5 was significantly up-regulated in tumor tissues compared with normal tissues (Figure 2B). Meanwhile, qRT-PCR was conducted to test KLF5 in CC cells, and the results showed that KLF5 mRNA was significantly up-regulated in CC cell lines relative to the normal cell line (Figure 2C).
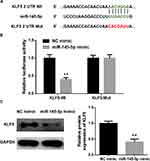
KLF5 Is a Direct Target of miR-145-5p in CC
As abovementioned, putative binding sites of miR-145-5p on KLF5 were predicted using the bioinformatics methods. To clarify the specific relationship between miR-145-5p and KLF5, NC mimic and miR-145-5p mimic were transfected into cancer cells. A dual-luciferase reporter assay was performed for further verification (Figure 3A and B) and revealed that overexpressing miR-145-5p could significantly decrease the luciferase activity of the cells transfected with Wt-KLF5, while there was no significant difference observed in the cells with Mut-KLF5. In addition, the protein level of KLF5 in HeLa cells transfected with miR-145-5p mimic was also inhibited (Figure 3C). These results collectively suggested that KLF5 was a direct target gene of miR-145-5p in CC.
Effects of the miR-145-5p/KLF5 Axis on Hela Cell Proliferation, Migration and Invasion
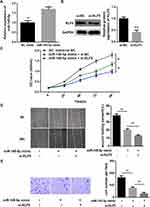
In the previous experiments, we verified that miR-145-5p could targeted regulate the expression of KLF5. In view of this, we further explored the role of the miR-145-5p/KLF5 regulatory axis in the malignant progression of CC. Hela cells were divided into three groups for in vitro experiments: NC mimic + si-NC, miR-145-5p mimic + si-NC and miR-145-5p mimic + si-KLF5 groups. qRT-PCR and Western blot were performed to determine the transfection efficiency, and found that miR-145-5p was significantly increased in the miR-145-5p mimic + si-NC group relative to the NC mimic + si-NC group, whereas KLF5 was remarkably decreased in the miR-145-5p mimic + si-KLF5 group relative to the miR-145-5p mimic + si-NC group (Figure 4A and B). Subsequently, cell biological behaviors were assayed. CCK-8, transwell invasion and wound healing assays suggested that miR-145-5p overexpression played an inhibitory role in cell proliferation, invasion and migration abilities, and such effect could be enhanced when KLF5 was simultaneously silenced (Figure 4C–E). Taken together, these results indicated that miR-145-5p could inhibit the proliferation, migration and invasion abilities of CC cells by down-regulating the expression of KLF5.
Discussion
Although the development of the HPV vaccine has contributed to the reduction of the risk of CC in women, it is still very important to explore the molecular mechanisms underlying the malignant progression of CC.23 Several studies have shown that miR-145 plays an anti-tumor role in a variety of human cancers.24,25 Sathyanarayanan et al found that miR-145 was down-regulated in CC, and the level of miR-145 showed an intimate correlation with prognosis.16,26 In the present study, miR-145-5p was found to be markedly decreased in CC tissues through the differential analysis on the miRNA expression data in the TCGA-CESC dataset. In addition, survival analysis showed that patients with high miR-145 had a better prognosis, which is in agreement with the previous studies.17,18 Moreover the expression of miR-145-5p in CC cell lines was also detected, and similar results were obtained.
At present, the direct target mRNAs of miR-145 in CC have been reported to be Fascin Actin-Bundling Protein 1 (FSCN1),27 SMAD-interacting protein 1 (SIP1),16 octamer-binding transcription factor 4 (OTC4)28 and myosin phosphatase targeting subunit 1 (MYTP1).29 In other words, KLF5 has not been studied as a target of miR-145-5p in CC. Although previous studies have shown that KLF5 is closely related to the occurrence and development of CC, the underlying molecular mechanism has not been fully discussed. Zhang et al reported that miR-152 functioned on CC by inhibiting KLF5.30 Besides, they found that KLF5 was highly expressed in CC and the down-regulation of KLF5 could result in the inhibition of cell proliferation and cell cycle progression but showed no relevance to the cell metastasis. Moreover, Ma et al suggested that KLF5 promoted the proliferation and invasion of CC cells, which partly depended on the expression of TNFRSF11a.31 Similarly, our study confirmed that KLF5 was highly expressed in CC. Furthermore, the results of in vitro experiments showed that miR-145-5p could inhibit the proliferation, migration and invasion of CC cells by down-regulating KLF5, which has not been reported in previous studies.32,33
To sum up, our study found that overexpressing miR-145-5p or silencing KLF5 could suppress the malignant progression of CC, and revealed the targeted relationship between the two genes for the first time. The discovery of the miR-145-5p/KLF5 regulatory axis also provides a theoretical support for mining molecular targets for CC targeted therapy. However, this study also has some limitations. For example, our research on the potential mechanism of miR-145-5p in the occurrence and development of CC has only focused on the target mRNA of miR-145-5p, which prompts us to make an in-depth study. In further study, we will analyze the enriched pathways of KLF5 in CC and explore the potential molecular mechanism of the miR-145-5p/KLF5-related signaling pathways underlying the malignant progression of CC.
Disclosure
The authors report no conflict of interest in this work.
References
1. Dizon DS, Mackay HJ, Thomas GM, et al. State of the science in cervical cancer: where we are today and where we need to go. Cancer. 2014;120(15):2282–2288. doi:10.1002/cncr.28722
2. Moore DH. Cervical cancer. Obstet Gynecol. 2006;107(5):1152–1161. doi:10.1097/01.AOG.0000215986.48590.79
3. Cervical cancer analysis reveals new mutations. Cancer Discov. 2017;7(4):344.
4. Forman D, de Martel C, Lacey CJ, et al. Global burden of human papillomavirus and related diseases. Vaccine. 2012;30(Suppl 5):F12–F23. doi:10.1016/j.vaccine.2012.07.055
5. Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16(1):1–17. doi:10.1128/CMR.16.1.1-17.2003
6. Schiffman M, Castle PE, Jeronimo J, et al. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890–907. doi:10.1016/S0140-6736(07)61416-0
7. Snijders PJ, Steenbergen RD, Heideman DA, et al. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208(2):152–164. doi:10.1002/(ISSN)1096-9896
8. Berindan-Neagoe I, Monroig PDC, Pasculli B, et al. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64(5):311–336. doi:10.3322/caac.21244
9. Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9(1):287–314. doi:10.1146/annurev-pathol-012513-104715
10. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
11. Hu X, Miao J, Zhang M, et al. miRNA-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing ATF7 in vitro. Mol Cells. 2018;41(5):390–400. doi:10.14348/molcells.2018.2078
12. Liu J, Bian T, Feng J, et al. miR-335 inhibited cell proliferation of lung cancer cells by target Tra2β. Cancer Sci. 2018;109(2):289–296. doi:10.1111/cas.13452
13. Wu J, Zhao Y, Li F, et al. MiR-144-3p: a novel tumor suppressor targeting MAPK6 in cervical cancer. J Physiol Biochem. 2019;75(2):143–152. doi:10.1007/s13105-019-00681-9
14. Li J, Chu Z-P, Han H, et al. Suppression of miR-93-5p inhibits high-risk HPV-positive cervical cancer progression via targeting of BTG3. Hum Cell. 2019;32(2):160–171. doi:10.1007/s13577-018-00225-1
15. Tang W, Zhang X, Tan W, et al. miR-145-5p suppresses breast cancer progression by inhibiting SOX2. J Surg Res. 2019;236:278–287. doi:10.1016/j.jss.2018.11.030
16. Sun M, Zhao W, Chen Z, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer. 2019;145(8):2170–2181. doi:10.1002/ijc.v145.8
17. Wei H, Wen-Ming C, Jun-Bo J. Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J Int Med Res. 2017;45(3):1054–1060. doi:10.1177/0300060517709614
18. Sathyanarayanan A, Chandrasekaran KS, Karunagaran D. microRNA-145 modulates epithelial-mesenchymal transition and suppresses proliferation, migration and invasion by targeting SIP1 in human cervical cancer cells. Cell Oncol (Dordr). 2017;40(2):119–131. doi:10.1007/s13402-016-0307-3
19. Huang L, Lin J-X, Yu Y-H, et al. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS One. 2012;7(3):e33762. doi:10.1371/journal.pone.0033762
20. Li MY, Hu XX. Meta-analysis of microRNA expression profiling studies in human cervical cancer. Med Oncol. 2015;32(6):510. doi:10.1007/s12032-015-0510-5
21. Ghaleb AM, NANDAN MO, CHANCHEVALAP S, et al. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15(2):92–96. doi:10.1038/sj.cr.7290271
22. Shi M, Du L, Liu D, et al. Glucocorticoid regulation of a novel HPV-E6-p53-miR-145 pathway modulates invasion and therapy resistance of cervical cancer cells. J Pathol. 2012;228(2):148–157. doi:10.1002/path.v228.2
23. Gao Y, Ding Y, Chen H, et al. Targeting Krüppel-like factor 5 (KLF5) for cancer therapy. Curr Top Med Chem. 2015;15(8):699–713. doi:10.2174/1568026615666150302105052
24. Shindo T, Manabe I, Fukushima Y, et al. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8(8):856–863. doi:10.1038/nm738
25. Joura EA, Giuliano AR, Iversen O-E, et al. A 9-valent HPV vaccine against infection and intraepithelial neoplasia in women. N Engl J Med. 2015;372(8):711–723. doi:10.1056/NEJMoa1405044
26. Boufraqech M, Zhang L, Jain M, et al. miR-145 suppresses thyroid cancer growth and metastasis and targets AKT3. Endocr Relat Cancer. 2014;21(4):517–531. doi:10.1530/ERC-14-0077
27. Lei C, Du F, Sun L, et al. miR-143 and miR-145 inhibit gastric cancer cell migration and metastasis by suppressing MYO6. Cell Death Dis. 2017;8(10):e3101. doi:10.1038/cddis.2017.493
28. Ye C, Sun N-X, Ma Y, et al. MicroRNA-145 contributes to enhancing radiosensitivity of cervical cancer cells. FEBS Lett. 2015;589(6):702–709. doi:10.1016/j.febslet.2015.01.037
29. Ma L, Li LL. miR-145 contributes to the progression of cervical carcinoma by directly regulating FSCN1. Cell Transplant. 2019;28(9–10):1299–1305. doi:10.1177/0963689719861063
30. Yan S, Li X, Jin Q, et al. MicroRNA-145 sensitizes cervical cancer cells to low-dose irradiation by downregulating OCT4 expression. Exp Ther Med. 2016;12(5):3130–3136. doi:10.3892/etm.2016.3731
31. González-Torres A, Bañuelos-Villegas EG, Martínez-Acuña N, et al. MYPT1 is targeted by miR-145 inhibiting viability, migration and invasion in 2D and 3D HeLa cultures. Biochem Biophys Res Commun. 2018;507(1–4):348–354. doi:10.1016/j.bbrc.2018.11.039
32. Zhang H, Lu Y, Wang S, et al. MicroRNA-152 acts as a tumor suppressor microRNA by inhibiting Krüppel-like factor 5 in human cervical cancer. Oncol Res. 2019;27(3):335–340. doi:10.3727/096504018X15252202178408
33. Ma D, Chang L-Y, Zhao S, et al. KLF5 promotes cervical cancer proliferation, migration and invasion in a manner partly dependent on TNFRSF11a expression. Sci Rep. 2017;7(1):15683. doi:10.1038/s41598-017-15979-1
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.