Back to Journals » Neuropsychiatric Disease and Treatment » Volume 16
miR-138 Increases Depressive-Like Behaviors by Targeting SIRT1 in Hippocampus
Authors Li C, Wang F, Miao P, Yan L, Liu S, Wang X, Jin Z, Gu Z
Received 6 November 2019
Accepted for publication 22 February 2020
Published 9 April 2020 Volume 2020:16 Pages 949—957
DOI https://doi.org/10.2147/NDT.S237558
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Cuixia Li,1 Feng Wang,2 Pei Miao,1 Libo Yan,1 Silin Liu,1 Xian Wang,1 Zuolin Jin,1 Zexu Gu1
1State Key Laboratory of Military Stomatology and National Clinical Research Center for Oral Diseases and Shaanxi Clinical Research Center for Oral Diseases, Department of Orthodontics, School of Stomatology, the Fourth Military Medical University, Xian 710032, People’s Republic of China; 2Department of Stomatology, 546 Hospital of PLA, Malan City, Xinjiang Province 841200, People’s Republic of China
Correspondence: Zuolin Jin; Zexu Gu
State Key Laboratory of Military Stomatology and National Clinical Research Center for Oral Diseases and Shaanxi Clinical Research Center for Oral Diseases, Department of Orthodontics, School of Stomatology, The Fourth Military Medical University, Xian 710032, People’s Republic of China
Email [email protected]; [email protected]
Background: Major depressive disorder (MDD) is a serious and common mood disorder with unknown etiology. Emerging evidence has demonstrated the critical roles of SIRT1 and microRNAs (miRNAs) in the progression of MDD. However, the underlying molecular mechanisms remain to be fully understood.
Methods: In the present study, the expression level of miR-138 and SIRT1 were analyzed by RT-PCR or Western blotting in a chronic unpredictable mild stress (CUMS) model. The depressive-like behaviors were analyzed by forced swimming test (FST) and sucrose preference test (SPT) in mice injected with miR-138 and SIRT1 overexpression lentivirus. The luciferase reporter assay was used to assess the direct regulation of miR-138 on SIRT1 expression.
Results: The upregulation of miR-138 was found in the hippocampus of the CUMS mice and correlated with decreased SIRT1 expression. C57BL/6J mice treated with SIRT1- and miR-138-expressing (miR-138) lentivirus showed increased depressive-like behaviors. In contrast, SIRT1 or si-miR-138 lentivirus treated C57BL/6J mice showed decreased depressive-like behaviors. Moreover, the Sirt1/PGC-1α/FNDC5/BDNF pathway was downregulated following miR-138 overexpression and increased upon miR-138 knockdown in hippocampus in CUMS mice and cultured primary neuronal cells. Mechanistically, luciferase reporter assay demonstrated that SIRT1 gene was a downstream transcriptional target of miR-138.
Conclusion: Our data demonstrated the regulation role of miR-138 on SIRT1 gene expression, miR-138 increased depressive-like behaviors by regulating SIRT1 expression in hippocampus.
Keywords: depression, microRNA, miR-138, SIRT1, chronic unpredictable mild stress, CUMS
Introduction
Major depressive disorder (MDD), or clinical depression, is a common mental disease that affects more than 300 million people worldwide according to World Health Organization’s report in 2018. It is estimated that major depression will rank as the first cause of burden of disease worldwide by 2030.1 Patients with depressive disorders often show functional impairments, which lead to social and occupational impairments that will disrupt their work, school, leisure, family life activities and responsibilities.2,3 Although substantial studies in genetic, molecular and neuroimaging studies have advanced our knowledge of the development and pathology of depression, the precise mechanism underlying depression is not clearly understood. Many different animal models have been established to study MDD, including chronic social defeat stress (CSDS),4 social separation,5 maternal separation stress (MSS),6 and chronic unpredictable mild stress (CUMS).7 Compared to other models, CUMS can better mimic the unpredictable intermittent stress exposure and nature of mild stress experience in humans. Thus, CUMS model is the most frequently used and widely accepted animal model for depressive-like behaviors.7
Non-coding RNAs are emerging as mega epigenetic mechanism of gene expression regulation and have been found to have critical function in the development for many diseases. Small noncoding RNAs can modulate gene expression through RNA-RNA interaction, RNA modification, RNA interference, and alternative splicing.8,9 Among small noncoding RNAs, microRNAs are the most well studied and characterized, they have been shown to regulate nearly 60% of all human genes and are involved in almost all biological functions.10 Numerous studies have revealed the crucial roles of microRNAs in regulating gene expression during the development of brain, synaptic plasticity as well as neuroplasticity and neurogenesis.11,12 Significant research showed that the dysregulation of miRNAs is involved in different degenerative diseases including Down syndrome, Alzheimer’s disease and Parkinson’s disease. miRNAs are also extensively involved in depression and stress-related diseases based on evidence from preclinical and clinical studies.13,14 Recent studies have compared the expression of miRNA in rats who developed stress-induced depression and those who did not. They found that depressive rats showed a robust adaptive miRNA when exposed to inescapable shocks, however, those rats that did not develop depression showed very limited miRNA expression change.15 Li et al demonstrated that CUMS could trigger upregulation of miR-182 in the hippocampus, overexpression of miR-182 increased depression-like behavior by downregulating the expression of BDNF.16 miR-138 expression was found to be upregulated by chronic restraint stress in male rats, further study showed miR-138 overexpression reduced the size of dendritic spines in cultured hippocampal neurons.17,18 However, the mechanism whereby miR-138 regulates neuron function and its role in depression are not clearly understood.
In our present study, we have shown evidence to demonstrate that miR-138 expression level was upregulated in the hippocampus of a CUMS model. Further luciferase reporter study confirmed that SIRT1 gene had miR-138 binding sites and was a downstream gene of miR-138 in hippocampus. Our findings are consistent with a previous study on miR-138, indicating critical roles of miR-138/SIRT1 axis in the development and pathology of depressive disorders.
Materials and Methods
Animals
All protocols were reviewed and approved by the Institute for Experimental Animals of School of Stomatology, Air Force Medical University. All the animal experiments followed the guidelines from IACUC (Institutional Animal Care and Use Committee). C57BL/6J mice were obtained from the Beijing Vital River Laboratory, China. All mice were housed in standard cages with a regular 12 h light and 12 h dark cycle at 22 ± 2°C and relative humidity at 55 ± 5%. The mice had unlimited access to chow diet and drinking water throughout the whole period of the experiment. All the mice were kept under the housing conditions for 7 days before behavior experiments.
Chronic Unpredictable Mild Stress (CUMS) Procedure
CUMS procedure was carried out as previously described.19 Briefly, the whole procedure lasted for 5 weeks including multiple different randomly assigned stressors as follows: a) the mice were forced to swim at 4°C for 5 min followed by 10 min dry-heat stress at 45°C; b) 30 min cage vibration and then constraint for 2 h; c) cage was inclined at 45° for 12 h and damp bedding for 16 h; d) 24 h continuous illumination; e) 24 h deprivation of drinking water and chow-diet. All animals were kept separately for CUMS stimulation, each animal received one kind of stressor per day and the single stressor was not conducted for two consecutive days.
Lentivirus Vectors
SIRT1 lentiviral vector (LV-SIRT1) construction and subsequent lentivirus packaging were completed by Gene Pharma Company (Shanghai, China). Lentivirus of miR-138 (107 TU/mL) and si-miR-138 (107 TU/mL) were also constructed and packaged in the Gene Pharma Company (Shanghai, China).
Stereotaxic Surgery for Injection of Control, SIRT1 and miR-138 Overexpression or Si-miR-138 Lentivirus
All mice subjected to surgery were anesthetized using K-X (100 and 10 mg/kg) by intraperitoneal injection and mounted onto a stereotaxic instrument. Bilateral craniotomies were conducted using a 0.5 mm diameter drill bit at –2.2 mm anteroposterior (±2.0 mm ML, +1.5 mm AP, −2.0 mm DV from bregma). The needle was positioned at target site for 2 min before injection. Then a total of 1 µL of Control, SIRT1 and miR-138 overexpression or si-miR-138 lentivirus was slowly injected into hippocampus using a glass micropipette attached to stereotaxic instrument, the needle was kept at the injection site for another 4 minutes after all of the virus was delivered. The mice were removed from stereotaxic instrument and placed into a warm cage for recovery. All the behavioral experiments were preformed one week after stereotaxic surgery.
Sucrose Preference Test (SPT)
The mice were single housed and kept in proper cages with two bottles of 1% (w/v) sucrose solution for 24 hours before the test. After that, mice underwent water and food deprivation for 24 hours. Then the mice were housed individually in cages with 1 bottle of water and 1 bottle of 1% (w/v) sucrose solution with free access. The volumes of remaining sucrose and water were determined 24 hours later, sucrose preference index (%) was measured using the following method:
sucrose preference index (%) = sucrose consumption/(water consumption + sucrose consumption) × 100.
Forced Swim Test (FST)
The FST was conducted 24 h post-SPT. Mice were placed into a plastic tank filled with tap water at 23–27°C for a period of 6 min (360 s), their behavior during the test was digitally recorded. Mobility time was judged when mice were making movements not only for keeping its head above water. The immobility time and mobility time were recorded throughout the 6 min observation time.
Elevated Plus-Maze (EPM)
The EPM was conducted 24 h post-FST. Mice were placed in the central area of an elevated plus-maze with a pair of open arms and closed arms, with their heads facing the open arms, after which each mouse was placed in the same position. At the same time, the camera monitor was opened to record the time spent in the open and closed arms and the total distance moved in open arm of the experimental animals within 5 min. During the experiment, the experimenter needs to be 1 m away from the maze. After the recording was finished, the experimental animals were returned to the breeding cage. The maze was cleaned and wiped with 5% acetic acid solution or 75% alcohol to eliminate the effect of animal odor on subsequent experimental animals.
Primary Cortical Neuron Culture
Cerebra cortices were isolated from mouse embryo brain and then dissociated with trypsin (Gibco) digestion to get single cell suspension. The single cells were spun down and re-suspended in a neurobasal medium (Gibco) supplemented with 2 mM GlutaMAX at the concentration of 1×105 cells/L. 300 cells were seeded in 35 mm cell culture dish and incubated in 5% CO2, 37°C incubator for 2 hrs. The medium was replaced with 3 mL of neurobasal/B27 medium and culturing was continued in 5% CO2 and 21% O2 conditions for 2 weeks. Then the cells were infected with miR-138 overexpression or knockdown lentivirus, the protein level of PGC-1α, FNDC5, BDNF, and SIRT1 were analyzed 48h later by immunoblotting.
Quantitative RT-PCR (qRT-PCR)
The total RNA was extracted in isolated hippocampus using TRIzol follow manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) assay was used to determine the mRNA expression level of target genes using the SYBR Premix Ex TaqTM II (Takara Co., Ltd., Dalian, China). The relative mRNA levels of each gene were calculated using the 2-ΔΔCT method. Primers for each gene were as follows: miR-138 forward: 5′- GACCCAGATTCCACCATAT −3′; reverse: 5′- CAGTGCAGGGTCCGAGGT −3′. SIRT1 forward: 5′- TGATTGGCACCGATCCTCG −3′; reverse: 5′- CCACAGCGTCATATCATCCAG −3′. U6 forward: 5′- CGCTTCGGCAGCACATATAC −3′; reverse: 5′- AAATATGGAACGCTTCACGA −3′. GAPDH forward: 5′- CATCACTGCCACCCAGAAGACTG −3′, reverse: 5′- ATGCCAGTGAGCTTCCCGTTCAG −3′. U6 snRNA and GAPDH were used here as internal control for miRNAs and genes, respectively.
Western Blotting
30 µg of total protein extracted from mouse brains or cell lysate was subjected to SDS-PAGE, separated proteins were transferred to nitrocellulose membrane and blocked in 5% BSA in TBS/0.1% Tween 20 (TBST) and then incubated with indicated primary antibodies: Sirt1 (Cell Signaling Technology, 1:1000), BDNF (Abcam, 1:1000), GAPDH (Abcam, 1:10000), PGC-1α (Cell Signaling Technology, 1:1000) and FNDC5 (Abcam, 1:1000) at 4 degrees overnight. The expression of indicated proteins was detected by incubating the blots with HPR-conjugated secondary antibodies.
Luciferase Reporter Assay
The potential binding sites of miR-138 within the SIRT1 3′-untranslated region (3′-UTR) (WT-SIRT1) or mutants (Mut-SIRT1) were inserted into the pmirGLO Dual-Luciferase miRNA target expression vector (Promega, Madison, WI). The luciferase reporter constructs were transfected into primary neuron cells, luciferase assays were performed using the Dual-Luciferase Reporter 1000 Assay System (Promega) according to manufacturer’s instructions. Data were normalized based on control Renilla luciferase signal.
Statistical Analysis
All statistical analyses were done by using GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA). Quantitative results are shown as mean ± SD, the difference was calculated using Student's t-tests and one-way analysis of variance (ANOVA), the level of significance was set at 0.05.
Sequence and time interval of each experiment and the position of the needles in hippocampus are shown in Figure S1.
Results
CUMS Induced the Expression of miR-138 and Decreased SIRT1 Expression
Rodent chronic unpredictable mild stress (CUMS) models, which replicate the specific neuroendocrinological and cytokine expression abnormalities of depression patients, are widely used to mimic depressive-like behaviors in humans. Here we examined the expression of miR-138 and SIRT1 in hippocampus by RT-PCR in C57BL/6J mice exposed to CUMS. Compared to control mice, miR-138 was significantly upregulated in mice subjected to CUMS (Figure 1A), conversely, the mRNA level of SIRT1 gene was decreased in CUMS mice (Figure 1B). Correlation analysis results showed miR-138 mRNA reversely correlates with SIRT1 expression (Figure 1C), indicating that miR-138 might regulate SIRT1 gene expression in the hippocampus of CUMS mice.
miR-138 Overexpression Induced Depressive-Like Behaviors
To demonstrate the potential function of miR-138 in CUMS exposed animals, C57BL/6J mice were introduced with LV-scramble, LV-miR-138 and LV-si-miR-138 lentivirus by stereotaxic injection, the depression-like behaviors were measured by sucrose preference test (SPT) and forced swimming test (FST). As shown in Figure 2, animals injected with miR-138 displayed a significant reduction in sucrose preference and increment of immobility time in FST test. In contrast, knockdown of miR-138 by LV-si-miR-138 lentivirus injection increased sucrose and mobility in FST test in animals exposed to CUMS. Moreover, in EPM experiment, the time LV-miR-138 group spent in open arm was significantly less than LV-scramble group, and the total distance moved in open arm did not show significant difference (Figure S2). Taken together, the behavior tests' results suggested that miR-138 induced depressive-like behaviors.
miR-138 Down-Regulates the Expression of Sirt1/PGC-1α/FNDC5/BDNF
Next, we sought to determine the underlying mechanism by which miR-138 increased depressive-like behaviors. The hippocampus of mice treated with LV-scramble, LV-miR-138 and LV-si-miR-138 lentivirus were isolated, the expression of genes of interest was measured by RT-PCR and Western blotting. The expression of miR-138 was significantly increased in LV-miR-138 injected mice while decreased in LV-si-miR-138 injected mice, confirming the efficiency of stereotaxic injection (Figure 3A). The mRNA level of Sirt1, PGC-1α, FNDC5 and BDNF were accordingly decreased in LV-miR-138 injected mice and increased in LV-si-miR-138 injected ones (Figure 3B–E). The protein expression level of Sirt1, PGC-1α, FNDC5 and BDNF were further confirmed by Western blotting using specific antibodies with GAPDH as an internal control (Figure 3F). These data demonstrated that miR-138 regulates depression-like behaviors through modulating Sirt1/PGC-1α/FNDC5/BDNF pathway.
SIRT1 Is a Direct Downstream Gene of miR-138
MicoRNAs regulate gene expression by directly binding to complementary DNA sequence with promoter regions. To investigate whether SIRT1 is the direct target of miR-138 in hippocampus, DNA sequence of SIRT1 gene was analyzed. Indeed, specific binding sites for miR-138 were found with 3ʹUTR of SIRT1 gene (Figure 4A). Luciferase reporter assays were performed in primary neuron cells to determine the regulation of miR-138 on the transcription of SIRT1. MiR-138 decreased the luciferase signal of wild-type (WT) SIRT1 3′UTR reporter gene but not the mutant (MUT) one (Figure 4B), indicating that miR-138 directly binds to the binding sites of 3ʹUTR of SIRT1 gene to regulate its transcription. Treatment of miR-138 decreased SIRT1 expression in primary neurons at both mRNA and protein level, knockdown of miR-138 with si-miR-138 reversely increased the expression of SIRT1 in primary neurons (Figure 4C and D). These data together suggested SIRT1 is a downstream gene for miR-138.
SIRT1 Overexpression Reduces Sucrose Consumption and Increases Mobility in CUMS Mice
To further elucidate the function of SIRT in depressive-like behavior induced by CUMS, SIRT1 was overexpressed in C57BL/6J mice by lentivirus injection, depressive-like behavior in mice was measured by SPT and FST tests. As shown in Figure 5, reduction of sucrose consumption and increase of mobility were observed in SIRT1 overexpressed mice, suggesting SIRT1 attenuates CUMS-induced depression-like behavior in rodent CUMS models.
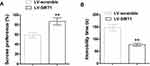 |
Figure 5 SIRT1 overexpression reduced CUMS-induced depression-like behavior. (A) Sucrose preference and (B) immobility time in FST of the mice. ** P < 0.01 compared with LV-scramble group. |
Discussion
As a highly expressed neurotrophic factor, brain-derived neurotrophic factor (BDNF) is involved in many aspects of the development of the brain.20 BNDF is essential for hippocampal function and synaptic plasticity. Val66Met mutation in BDNF gene has been found to cause decrease of BDNF secretion and increased anxiety and depression, decreased BDNF level is correlated with the severity and recurrence of depression.21,22 In this study, we found that miR-138 upregulation increased depressive-like behavior in CUMS induced mice, further study showed SIRT1 is a downstream gene of miR-138. miR-138 decreased expression level of BDNF through SIRT1/PGC-1α/FNDC5/BDNF pathway in cultured neuron. A previous study also showed miR-138 expression was increased by chronic restraint stress. Our data combined with previous study demonstrate a potential role of miR-138 in the pathology of depression.
Sirtuins proteins are nicotinamide adenine dinucleotide (NAD+) dependent deacylases found in all aerobic organisms, they are involved in many different biological processes, including aging, metabolic pathway, cell differentiation, DNA damage repair, apoptosis and inflammation.23–25 Sirt1 protein is composed of 747 amino acids, with a conserved deacetylase domain and very flexible N- and C- terminals, providing multiple modulation sites for post-translational modifications and interactions with other proteins.26 Recent studies have revealed an essential role of Sirt1 in drug addiction, endocrine regulation and synaptic plasticity induced brain dysfunctions.27,28 Several studies showed that dysregulation of Sirt1 gene and expression level are associated with depression. A case-control study in Japan showed that SNP (rs10997875) in the Sirt1 gene may be associated with depression. The level of Sirt1 in peripheral blood is decreased in patients with depression compared to normal individuals.29–31 Dysregulation of Sirt1 expression was shown to be related to depressive-like behaviors in animal model of depression, activation of Sirt1 blocked chronic stress induced depression-like phenotypes, in contrast, genetic ablation or pharmacological inhibition of Sirt1 in hippocampus increased depression-like behaviors.32
Peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α) is a transcriptional co-activator that has important functions in mitochondrial biogenesis and oxidative response pathway.33 Many studies have shown important roles of PGC-1a in brain function. PGC-1a deficiency is associated with neurodegeneration and increased risk of Parkinson’s disease in the MPTP mouse model.34,35 PGC-1α is also involved in the formation and maintenance of neuronal dendritic spines by regulation of BNDF expression.36 FNDC5 is a PGC-1α-dependent myokine which plays important roles in neuron development; lack of FNDC5 impaired the development of neuronal precursors into mature neurons.37,38 A study by Wrann et al showed that endurance exercise could induce BNDF in hippocampus through a PGC-1α/FNDC5 pathway.39 The activity of PGC-1α is highly regulated by its acetylation status, mutation of PGC-1α acetylation sites significantly increases the transcriptional activity of PGC-1α protein. Interestingly, studies have demonstrated that SIRT1 can directly interact with PGC-1α and mediate its deacetylation, and induction of SIRT1 by fasting and dietary restriction upregulates PGC-1α activity.39,40 Zhao et al also found that high-fat diet exacerbated isoflurane-induced postoperative cognitive dysfunction through suppression of the expression of the Sirt1/PGC-1α/FNDC5/BDNF pathway in the hippocampus.41 Our study has identified that SIRT1 is a downstream gene of miR-138, overexpression of miR-138 significantly downregulated SIRT1 expression level as well as PGC-1α, FNDC5 and BDNF expression, suggesting miR-138’s role in depression is through regulation of Sirt1/PGC-1α/FNDC5/BDNF pathway.
Emerging studies have demonstrated the important roles of miRNAs in many aspects of neurogenesis, neural plasticity and stress response, strong evidence also indicated the critical roles of miRNAs in depression.42 Besides miR-138 that has been reported here, miR-155 knockout mice showed reduced anxiety and depression-like behaviors in forced swim test.43 Studies also suggest miR-124 could serve as a putative therapeutic target and biomarker for major depression.44 It has also been found that depression patients had lower miR-135a in their blood and its level was increased after antidepressant treatment.45 Research regarding miRNAs and depression will provide more insight into using miRNA as a biomarker for the pathology and find potential novel therapeutic strategies for depression. Taken together, our study provided evidence that miR-138 increased depressive-like behaviors through regulating SIRT1 expression and BDNF level, further study will be granted to focus on the level of miR-138 in peripheral blood in animal models and depression patients to explore miR-138 as a biomarker or therapeutic target for depression.
Ethics and Consent Statement
The present study was approved by the Ethics Committee of the Fourth Military Medical University.
Funding
This work was supported by the National Nature Science Foundation of China (Nos. 81571000 and 81970960) and Natural Science Foundation of Shaanxi Province (No2017jz030).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Malhi GS, Mann JJ. Depression. Lancet. 2018;392(10161):2299–2312. doi:10.1016/S0140-6736(18)31948-2
2. Kupfer DJ, Frank E, Phillips ML. Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet. 2012;379(9820):1045–1055. doi:10.1016/S0140-6736(11)60602-8
3. Sheehan DV, Nakagome K, Asami Y, Pappadopulos EA, Boucher M. Restoring function in major depressive disorder: a systematic review. J Affect Disord. 2017;215:299–313. doi:10.1016/j.jad.2017.02.029
4. Huang GB, Zhao T, Muna SS, et al. Effects of chronic social defeat stress on behaviour, endoplasmic reticulum proteins and choline acetyltransferase in adolescent mice. Int J Neuropsychopharmacol. 2013;16(7):1635–1647. doi:10.1017/S1461145713000060
5. Grippo AJ, Gerena D, Huang J, et al. Social isolation induces behavioral and neuroendocrine disturbances relevant to depression in female and male prairie voles. Psychoneuroendocrinology. 2007;32(8–10):966–980. doi:10.1016/j.psyneuen.2007.07.004
6. Matthews K, Robbins TW. Early experience as a determinant of adult behavioural responses to reward: the effects of repeated maternal separation in the rat. Neurosci Biobehav Rev. 2003;27(1–2):45–55.
7. Biala G, Pekala K, Boguszewska-Czubara A, et al. Behavioral and biochemical impact of chronic unpredictable mild stress on the acquisition of nicotine conditioned place preference in rats. Mol Neurobiol. 2018;55(4):3270–3289. doi:10.1007/s12035-017-0585-4
8. D’Sa C, Duman RS. Antidepressants and neuroplasticity. Bipolar Disord. 2002;4(3):183–194. doi:10.1034/j.1399-5618.2002.01203.x
9. Schouten M, Buijink MR, Lucassen PJ, Fitzsimons CP. New neurons in aging brains: molecular control by small non-coding RNAs. Front Neurosci. 2012;6:25. doi:10.3389/fnins.2012.00025
10. Malphettes L, Fussenegger M. Impact of RNA interference on gene networks. Metab Eng. 2006;8(6):672–683. doi:10.1016/j.ymben.2006.07.005
11. Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35(5):325–334. doi:10.1016/j.tins.2012.01.004
12. Fries GR, Zhang W, Benevenuto D, Quevedo J. MicroRNAs in major depressive disorder. Adv Exp Med Biol. 2019;1118:175–190.
13. Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity–a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014;369(1652). doi:10.1098/rstb.2013.0515
14. Hollins SL, Cairns MJ. MicroRNA: small RNA mediators of the brains genomic response to environmental stress. Prog Neurobiol. 2016;143:61–81. doi:10.1016/j.pneurobio.2016.06.005
15. Munoz-Llanos M, Garcia-Perez MA, Xu X, et al. MicroRNA profiling and bioinformatics target analysis in dorsal hippocampus of chronically stressed rats: relevance to depression pathophysiology. Front Mol Neurosci. 2018;11:251. doi:10.3389/fnmol.2018.00251
16. Li Y, Li S, Yan J, et al. miR-182 (microRNA-182) suppression in the hippocampus evokes antidepressant-like effects in rats. Prog Neuropsychopharmacol Biol Psychiatry. 2016;65:96–103. doi:10.1016/j.pnpbp.2015.09.004
17. Siegel G, Obernosterer G, Fiore R, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11(6):705–716. doi:10.1038/ncb1876
18. Castaneda P, Munoz M, Garcia-Rojo G, et al. Association of N-cadherin levels and downstream effectors of Rho GTPases with dendritic spine loss induced by chronic stress in rat hippocampal neurons. J Neurosci Res. 2015;93(10):1476–1491. doi:10.1002/jnr.23602
19. Wang Y, Xu J, Liu Y, Li Z, Li X. TLR4-NF-kappaB signal involved in depressive-like behaviors and cytokine expression of frontal cortex and hippocampus in stressed C57BL/6 and ob/ob mice. Neural Plast. 2018;2018:7254016. doi:10.1155/2018/7254016
20. Cotman CW, Berchtold NC, Christie LA. Exercise builds brain health: key roles of growth factor cascades and inflammation. Trends Neurosci. 2007;30(9):464–472. doi:10.1016/j.tins.2007.06.011
21. Greenberg ME, Xu B, Lu B, Hempstead BL. New insights in the biology of BDNF synthesis and release: implications in CNS function. J Neurosci. 2009;29(41):12764–12767. doi:10.1523/JNEUROSCI.3566-09.2009
22. Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci. 2013;14(1):7–23. doi:10.1038/nrn3379
23. Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75(1):435–465. doi:10.1146/annurev.biochem.74.082803.133500
24. Yang Y, Fu W, Chen J, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9(11):1253–1262. doi:10.1038/ncb1645
25. Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem Sci. 2008;33(11):517–525. doi:10.1016/j.tibs.2008.08.001
26. Huhtiniemi T, Wittekindt C, Laitinen T, et al. Comparative and pharmacophore model for deacetylase SIRT1. J Comput Aided Mol Des. 2006;20(9):589–599. doi:10.1007/s10822-006-9084-9
27. Gao J, Wang WY, Mao YW, et al. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466(7310):1105–1109. doi:10.1038/nature09271
28. Michan S, Li Y, Chou MM, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30(29):9695–9707. doi:10.1523/JNEUROSCI.0027-10.2010
29. Kishi T, Yoshimura R, Kitajima T, et al. SIRT1 gene is associated with major depressive disorder in the Japanese population. J Affect Disord. 2010;126(1–2):167–173. doi:10.1016/j.jad.2010.04.003
30. Kovanen L, Donner K, Partonen T, Li S. SIRT1 polymorphisms associate with seasonal weight variation, depressive disorders, and diastolic blood pressure in the general population. PLoS One. 2015;10(10):e0141001. doi:10.1371/journal.pone.0141001
31. Luo XJ, Zhang C. Down-regulation of SIRT1 gene expression in major depressive disorder. Am J Psychiatry. 2016;173(10):1046. doi:10.1176/appi.ajp.2016.16040394
32. Abe-Higuchi N, Uchida S, Yamagata H, et al. Hippocampal sirtuin 1 signaling mediates depression-like behavior. Biol Psychiatry. 2016;80(11):815–826. doi:10.1016/j.biopsych.2016.01.009
33. Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282(4):647–672. doi:10.1111/febs.13175
34. St-Pierre J, Drori S, Uldry M, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi:10.1016/j.cell.2006.09.024
35. Corona JC, Duchen MR. PPARgamma and PGC-1alpha as therapeutic targets in Parkinson’s. Neurochem Res. 2015;40(2):308–316. doi:10.1007/s11064-014-1377-0
36. Cheng A, Wan R, Yang JL, et al. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nat Commun. 2012;3(1):1250. doi:10.1038/ncomms2238
37. Bostrom P, Wu J, Jedrychowski MP, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481(7382):463–468. doi:10.1038/nature10777
38. Hashemi MS, Ghaedi K, Salamian A, et al. Fndc5 knockdown significantly decreased neural differentiation rate of mouse embryonic stem cells. Neuroscience. 2013;231:296–304. doi:10.1016/j.neuroscience.2012.11.041
39. Wrann CD, White JP, Salogiannnis J, et al. Exercise induces hippocampal BDNF through a PGC-1alpha/FNDC5 pathway. Cell Metab. 2013;18(5):649–659. doi:10.1016/j.cmet.2013.09.008
40. Coste A, Louet JF, Lagouge M, et al. The genetic ablation of SRC-3 protects against obesity and improves insulin sensitivity by reducing the acetylation of PGC-1{alpha}. Proc Natl Acad Sci U S A. 2008;105(44):17187–17192. doi:10.1073/pnas.0808207105
41. Zhao Z, Yao M, Wei L, Ge S. Obesity caused by a high-fat diet regulates the Sirt1/PGC-1alpha/FNDC5/BDNF pathway to exacerbate isoflurane-induced postoperative cognitive dysfunction in older mice. Nutr Neurosci. 2019;1–12. doi:10.1080/1028415X.2019.1581460
42. Dwivedi Y. Emerging role of microRNAs in major depressive disorder: diagnosis and therapeutic implications. Dialogues Clin Neurosci. 2014;16(1):43–61.
43. Fonken LK, Gaudet AD, Gaier KR, Nelson RJ, Popovich PG. MicroRNA-155 deletion reduces anxiety- and depressive-like behaviors in mice. Psychoneuroendocrinology. 2016;63:362–369. doi:10.1016/j.psyneuen.2015.10.019
44. Dwivedi Y. microRNA-124: a putative therapeutic target and biomarker for major depression. Expert Opin Ther Targets. 2017;21(7):653–656. doi:10.1080/14728222.2017.1328501
45. Issler O, Haramati S, Paul ED, et al. MicroRNA 135 is essential for chronic stress resiliency, antidepressant efficacy, and intact serotonergic activity. Neuron. 2014;83(2):344–360. doi:10.1016/j.neuron.2014.05.042
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.