Back to Journals » OncoTargets and Therapy » Volume 13
miR-135b-5p Suppresses Androgen Receptor-Enhanced Hepatocellular Carcinoma Cell Proliferation via Regulating the HIF-2α/c-Myc/P27 Signals in vitro
Authors Bao S, Wang C, Jin S, Hu K, Lu J
Received 28 June 2020
Accepted for publication 18 September 2020
Published 7 October 2020 Volume 2020:13 Pages 9991—10000
DOI https://doi.org/10.2147/OTT.S268214
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Shi-xiang Bao,1,* Chun-hua Wang,2,* Shuai Jin,1 Kong-wang Hu,2 Jing-tao Lu1,3
1School of Life Sciences, Anhui Medical University, Hefei 230032, People’s Republic of China; 2Departments of General Surgery, The First Affiliated Hospital, Anhui Medical University, Hefei 230032, People’s Republic of China; 3George Whipple Laboratory for Cancer Research, Departments of Pathology and Urology and the Wilmot Cancer Center, University of Rochester Medical Center, Rochester, NY 14642, USA
*These authors contributed equally to this work
Correspondence: Jing-tao Lu; Kong-wang Hu
Anhui Medical University, 81 Meishan Road, Shushan District, Hefei 230032, People’s Republic of China
Tel/Fax +86 551 65160393
Email [email protected]; [email protected]
Introduction: Hepatocellular carcinoma (HCC) accounts for more than 90% of liver cancers and is ranked as the fifth most common malignancy. Androgen receptor (AR) may promote the progression of HCC at an early stage of the disease. However, this study identified miR-135b-5p as an AR upstream regulator can suppress AR protein expression and inhibit HCC proliferation, consistent with the idea that AR expression is negatively correlated with HCC progression.
Methods: The target microRNAs were predicted using online databases (TargetScan, miRDB, and MicroCosm Targets). Cell proliferation ability was measured by MTT and colony formation assay. Western blot was performed to analyze the expression levels of AR, HIF-2α, c-Myc, and p27, which are related to HCC proliferation. Chromatin immunoprecipitation (ChIP) assay and luciferase reporter assay were carried out to investigate the mechanism by which miR-135b-5p decreases AR expression.
Results: miR-135b-5p suppresses HCC cell proliferation and AR expression. Downregulation of AR expression by miR-135b-5p may in turn transcriptionally modulate HIF-2α expression via direct binding of AR to the androgen response element (ARE) in the HIF-2α promoter. Further dissection of the mechanism revealed that AR-modulated HIF-2α could suppress c-Myc expression resulting in increased p27 expression that likely contributes to the suppression of proliferation in HCC cells.
Conclusion: miR-135b-5p suppresses HCC cell proliferation via targeting AR-modulated HIF-2α/c-Myc/p27 signals, which may help to develop more effective therapies to prevent HCC progression.
Keywords: HCC, miR-135b-5p, AR, HIF-2α, c-Myc, p27
Introduction
Hepatocellular carcinoa (HCC) accounts for more than 90% of liver cancers and is ranked as the fifth most common malignancy.1 It is the second leading cause of cancer-induced death in males and the sixth in females with more than 700,000 deaths every year worldwide.2 While targeted molecular therapy and surgical resection have experienced rapid progress recently, the advancement of long-term survival remains far from satisfactory. Moreover, patients who have received surgical resection often have a high recurrence, with a 5-year survival rate of only 25%.3,4 Therefore, new and better therapies are needed to defeat this deadly disease.
Androgen receptor (AR), a member of nuclear hormone receptors, plays an important role in HCC progression.5 It has been reported that AR might promote HCC progression in its early developmental stages,6–9 and targeting AR can inhibit HBV- or carcinogen-induced HCC development at an early stage in vitro and in vivo.8 However, the molecular mechanisms leading to the activation of AR signaling during HCC progression remain to be further elucidated.
microRNAs (miRNAs) are a group of small, well-conserved, non-coding, endogenous RNA molecules containing around 18–25 nucleotides,10 which play key post-transcriptional regulatory roles in mRNA translation and degradation, predominantly in the 3ʹ-untranslated region (3ʹ-UTR).11,12 Recent studies have indicated that aberrantly expressed miRNAs might play crucial roles in a variety of biological processes, including cancer pathogenesis.13,14 Certain miRNAs have been shown to regulate tumor progression via binding to the AR 3ʹ-UTR sequence.15–17 As the expression of miRNAs is associated with HCC progression and metastasis,18–20 and AR might promote HCC cell proliferation, it would be interesting to determine if miRNAs serve as AR upstream regulators to alter HCC cell proliferation. Bioinformatic analyses from literature and online databases (TargetScan, miRDB, and MicroCosm Targets) suggest that a subset of miRNAs, including miR-99b-5p, miR-101-3p, miR-135b-5p, miR-373-3p, and miR-375, may target the AR 3ʹ-UTR sequence and simultaneously inhibit HCC progression. Our experiments identified miR-135b-5p as an AR upstream regulator, which suppresses HCC cell proliferation via downregulating the AR-mediated HIF-2α/c-Myc/p27 signal pathway.
Materials and Methods
Materials
MISSION® Synthetic miR-135b-5p inhibitor was purchased from Sigma-Aldrich (St. Louis, MO, USA) and transfected according to the technical bulletin provided by the manufacturer. AR, HIF-2α, c-Myc, and GAPDH antibodies for Western blotting were purchased from Santa Cruz Biotechnology. The p27 antibody for Western blotting was purchased from Cell Signaling. Anti-mouse/rabbit secondary antibodies for Western blotting were from Invitrogen.
In vitro Cell Culture/Maintenance
SK-hep1, HepG2, SNK, and Huh7 human HCC cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). HA22T human HCC cell line was purchased from the Food Industry Research and Development Institute in Taiwan (BCRC number: 60,168). All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS), penicillin (25 U/mL), streptomycin (25 g/mL), and 1% (v/v) L-glutamine in a 5% (v/v) CO2 humidified incubator at 37°C.
Lentiviral-Based Gene Delivery
The pWPI/pWPI-AR, PLVTHM plasmid, and pLVTHM plasmids containing the miRNAs, psAX2 packaging plasmid, and pMD2G envelope plasmid were transfected into 293T cells using the standard calcium phosphate transfection method. The lentivirus soups were collected after incubating for 48 h and concentrated by density gradient centrifugation before freezing at −80°C for later use.
Cell Proliferation Assay
Cells were seeded in 24-well plates (1x104 cells/well) and cultured in a 5% (v/v) CO2 humidified incubator at 37°C. The following day, the lentivirus soups of pWPI/pWPI-AR, PLVTHM, and PLVTHM-miR-135b-5p were added for 6 h, and the cells were continually cultured for 48 h. MTT agent was added, and OD (570 nm) was determined.
Colony Formation Assay
Cells were seeded in 6-well plates (1x103 cells/well) with three repetitions. After 12-day incubation, these plates were washed with phosphate buffered saline (PBS) twice, fixed by methanol for 10 min and stained with 0.1% crystal violet solution within 10 min for further analysis.
RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR Analysis (qRT-PCR)
Total RNAs were isolated using TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. Reverse transcription was performed using 1 µg of total RNA and Superscript III Reverse Transcriptase (Invitrogen, Grand Island, NY, USA). Quantitative real-time PCR (qRT-PCR) used a Bio-Rad CFX96 system with SYBR green to determine the mRNA expression level of the gene of interest. Expression levels were normalized to the expression of GAPDH mRNA. The qRT-PCR protocol was as follows: 50°C for 2 min, 95°C for 8 min 30sec, followed by 45 cycles at 95°C for 15 s and 60°C for 1 min. The extension was 95°C for 1 min, 55°C for 1 min, and 55°C for 10 s.
The PureLink® miRNA kit was used to extract the miRNAs. Then, 2 µg of RNA was incubated with 1 U of poly (A) polymerase with 1 mM ATP in 1x RT buffer at 37°C for 15 min in a volume of 10 μL. Reverse transcription was performed with the addition of 50 µM RT anchor primer and annealing at 65°C for 5 min, then 4°C for 2 min. Finally, cDNA synthesis was performed at 42°C for 60 min after the addition of 2 μL 10 mM dNTPs, 2 μL 5x RT buffer, 1 μL reverse transcriptase, and ddH2O to a total volume of 20 μL. The qRT-PCR protocol was as follows: 95°C for 2 min, followed by 45 cycles at 95°C for 15 s, and 60°C for 45 s using a Bio-Rad CFX96 system with SYBR green to determine the mRNA expression level of the gene of interest. Expression levels were normalized to the expression of 5s RNA and/or U6.
Western Blot Analysis
Cells were lysed in lysis buffer, and proteins (30 µg) were separated on 10–12% gel by SDS/PAGE before transferring onto PVDF membranes (Millipore, Billerica, MA, USA). After blocking, the membranes were incubated with appropriate dilutions of specific primary antibodies against GAPDH, AR (Santa Cruz Biotechnology, Dallas, TX, USA), HIF-2α, c-Myc, or p27 (Cell Signaling). The blots were incubated with HRP-conjugated secondary antibodies (Invitrogen, USA) and visualized using the ECL system (Thermo Fisher Scientific, Rochester, NY, USA).
Luciferase Reporter Assay
AR 3ʹ-UTR fragments of 540 bp containing wild-type or mutant miRNA response elements (MREs) were cloned into the psiCHECK-2 vector (Promega, Madison, WI, USA) downstream of the Renilla luciferase ORF. Cells were plated in 24-well plates, and the cDNAs were transfected using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. Luciferase activity was measured by the Dual-Luciferase Assay (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Chromatin Immunoprecipitation Assay (ChIP)
Cells were crosslinked with 4% formaldehyde for 10 min, followed by cell collection and sonication with a predetermined power to yield genomic DNA fragments of about 400 bp in length. Lysates were pre-cleared sequentially with normal rabbit IgG (sc-2027, Santa Cruz Biotechnology, Dallas, TX, USA) and protein A-agarose. Anti-AR antibody (2.0 µg) was added to the cell lysates, which were then incubated at 4°C overnight. For the negative control, IgG was used in the reaction. The target sequence within the human HIF-2α promoter was amplified using the following primers: Forward 5ʹ-GGGCCGAGAAATTATCCCCA −3ʹ; Reverse 5ʹ-GTT TATGAGGTCGTCTGGGC-3ʹ. PCR products were analyzed by agarose gel electrophoresis.
Statistical Analysis
Data are expressed as mean ± SD from at least three independent experiments. Statistical analyses involved Student’s t-test with GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). P <0.05 was considered statistically significant.
Results
miR-135b-5p Suppresses HCC Cell Proliferation
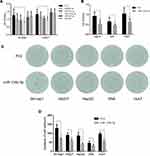
Since AR may promote HCC cell proliferation, an upstream regulator of AR may also be involved in this process. We identified a subset of miRNAs, including miR-99b-5p, miR-101-3p, miR-135b-5p, miR-373-3p, and miR-375, that are predicted to regulate AR expression while also being involved in HCC tumorigenesis. This hypothesis was directly tested by overexpressing these miRNAs in HCC cell lines. Cell proliferation assays showed that miR-135b-5p significantly suppressed HCC cell proliferation in SK-hep1 and HA22T cells (Figure 1A) and HepG2, SNK, and Huh7 cells (Figure 1B).
Importantly, a colony formation assay showed that miR-135b-5p significantly suppressed colony formation in HCC cell lines (Figure 1C and D), further supporting the role of miR-135b-5p as a suppressor of HCC progression.
miR-135b-5p Suppresses AR Protein Expression in HCC Cell Lines
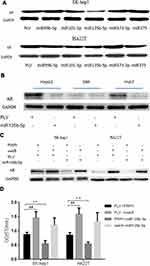
To link the suppression of HCC cell proliferation by miR-135b-5p to the modulation of AR expression, we first examined the effect of miR-135b-5p on AR expression in HCC cell lines. Analysis of AR expression by Western blot revealed that miR-135b-5p could significantly suppress the expression of AR protein in SK-hep1 and HA22T cells (Figure 2A) and HepG2 and Huh7 cells (Figure 2B).
To determine whether miR-135b-5p might function via modulation of AR expression to suppress HCC cell proliferation, we applied the interruption approach. The results revealed that over-expression of AR significantly increased AR protein expression in SK-hep1 and HA22T cells, and promoted their proliferation, while miR-135b-5p suppressed the expression of AR and decreased the proliferation of SK-hep1 and HA22T cells. However, in AR-overexpressing SK-hep1 and HA22T cells, miR-135b-5p did not suppress the expression of AR or decrease HCC cell proliferation (Figure 2C and D). This could be due to exogenous AR mRNA lacking the 3ʹ-UTR, thus the miR-135b-5p target sequence.
Together, the results suggest that miR-135b-5p may suppress HCC cell proliferation via modulation of AR expression.
Mechanism by Which miR-135b-5p Decreases AR Expression
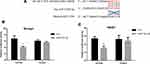
To further dissect the mechanism by which miR-135b-5p decreases AR expression at a molecular level, we first identified a potential MRE that might match the seed sequence of miR-135b-5p in the 3ʹ-UTR of AR gene. The MRE was mutated, and both wild-type and mutated MREs were cloned into the Renilla luciferase vector (Figure 3A). A luciferase reporter assay using SK-hep1 and HA22T cells revealed that miR-135b-5p decreased the relative luciferase activity with the wild-type AR 3ʹ-UTR, but there was no effect on the relative luciferase activity with the mutated AR 3ʹ-UTR (Figure 3B and C), suggesting that miR-135b-5p could directly target AR 3ʹ-UTR to suppress AR expression.
Downregulation of AR Expression by miR-135b-5p May Alter HIF-2α/c-Myc/P27 Signals to Suppress HCC Cell Proliferation
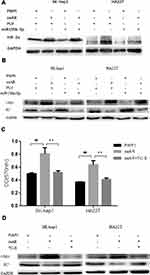
To dissect the mechanism by which suppression of AR expression by miR-135b-5p might alter HCC cell proliferation, we focused on HIF-2α/c-Myc/p27 signals. Recent studies, including in renal cell carcinoma, have indicated that AR might alter HIF-2α/c-Myc/p27 signals to promote tumor progression.21 Overexpression of AR in SK-hep1 and HA22T cells increased the expression of HIF-2α and c-Myc but decreased expression of p27. Expression of miR-135b-5p in SK-hep1 and HA22T cells resulted in decreased expression of HIF-2α and c-Myc and increased expression of p27. However, in AR-overexpressing SK-hep1 and HA22T cells, miR-135b-5p failed to suppress the increased expression of HIF-2α and c-Myc or to increase the decreased expression of p27 caused by overexpression of AR (Figure 4A and B).
To further confirm that miR-135b-5p suppresses HCC cell proliferation through regulation of AR-mediated HIF-2α/c-Myc/p27 signals, we applied the interruption approach adding TC-S, an inhibitor of HIF-2α. The results revealed that TC-S could suppress the levels of HCC cell proliferation observed in AR-overexpressing SK-hep1 and HA22T cells (Figure 4C). Importantly, TC-S could reverse the AR-modulated expression of c-Myc and p27 in SK-hep1 and HA22T cells (Figure 4D).
Together, these results suggest that miR-135b-5p suppresses HCC cell proliferation via altering AR-modulated HIF-2α/c-Myc/p27 signals in SK-hep1 and HA22T cells.
Mechanism by Which AR Modulates HIF-2α Expression at a Molecular Level in SK-hep1
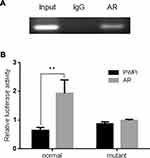
To further dissect the molecular mechanism by which AR can modulate HIF-2α expression in SK-hep1, bioinformatic analysis using Ensembl and PROMO 3.0 was performed to search for potential androgen response elements (AREs) in the HIF-2α promoter region. A potential ARE was found within 1.5 kb upstream of the transcriptional start site. Investigation by ChIP assay showed that AR could bind to the ARE located on the HIF-2α 5ʹ promoter (Figure 5A). A luciferase reporter assay using a construct containing the HIF-2α 5ʹ promoter sequence with either the wild-type or mutant ARE was performed in SK-hep1 cells. Increased luciferase activity was observed with the wild-type ARE but not with the mutant ARE (Figure 5B).
Together, these results suggest that AR can transcriptionally modulate HIF-2α expression via direct binding to the ARE located in the HIF-2α 5ʹ promoter region in HCC SK-hep1 cells.
Discussion
Recent studies have indicated that targeting AR could suppress HBV-22 HCV-,23 and carcinogen-24 induced HCC development at an early stage. Based on these discoveries, several clinical trials have been proposed to test the efficacy of anti-androgen therapy to suppress HCC progression. It was discovered that androgen deprivation therapy (ADT) using anti-androgens could significantly suppress HCC progression.25 However, Dai et al found that ADT failed to yield fruitful results.26 Instead of using anti-androgens to prevent androgen binding to AR, Ma et al27 and Wu et al28 applied the AR degradation enhancer ASC-J9 to directly target AR to suppress HCC progression. The experiments reported in this manuscript further support the notion that AR can increase the proliferation of HCC promoting its progression. We have also provided detailed molecular mechanisms of this process describing the roles of miR-135b-5p and AR target gene HIF2α.
Growing evidence indicates that the deregulation of miRNAs occurs in HCC progression29 and recent studies have further developed the miRNA-based risk of HCC recurrence following liver resection.30 Results of miRNA therapies in HCC showed that either reintroducing suppressor miRNAs or inhibiting oncogenic miRNAs suppress HCC progression in various HCC cell lines in vitro, animal models in vivo, and human clinical trials.31 Our work provides additional details, identifying a specific miRNA and its target gene, which are involved in the regulation of HCC proliferation. We first identified several miRNAs as potential AR upstream regulators including miR-99b-5p, miR-101-3p, miR-135b-5p, miR-373-3p, and miR-375. An MTT assay showed that the exogenous expression of miR-135b-5p was capable of suppressing cell proliferation in SK-hep1, HA22T, HepG2, SNK, and Huh7 HCC cell lines. The miRNAs miR-99b-5p and miR-101-3p also inhibited proliferation of the HCC cell lines SK-hep1 and HA22T, however, the level of inhibition was not statistically significant. In addition, miR-135b-5p also inhibited colony formation in SK-hep1, HA22T, HepG2, SNK, and Huh7 HCC lines. It is likely that the target of miR-135b-5p in HCC is AR since miR-135b-5p significantly suppressed the expression of AR in HCC cell lines. Furthermore, the luciferase reporter assay using SK-hep1 and HA22T cells showed that miR-135b-5p decreased the relative luciferase activity with the wild-type AR 3ʹ-UTR but not with the mutant AR 3ʹ-UTR. Moreover, miR-135b-5p failed to suppress AR expression and HCC cell proliferation in AR-overexpressing SK-hep1 and HA22T cells since exogenously expressed AR lacks the 3ʹ-UTR sequence.
When characterizing the molecules involved in regulating HCC proliferation downstream of miR-135b-5p-regulated AR expression, we found that HIF-2α/c-Myc/p27 signals likely play a significant role, similar to the findings in lung cancer and renal cell carcinoma (RCC).21 Overexpression of AR increased expression of HIF-2α and c-Myc but decreased expression of p27 in HCC. The addition of miR-135b-5p suppressed the expression of HIF-2α and c-Myc but increased the expression of p27. However, miR-135b-5p did not suppress the increased expression of HIF-2α and c-Myc or increase the decreased expression of p27 in AR-overexpressing HCC cells. The HIF-2α inhibitor TC-S suppressed the levels of HCC cell proliferation observed in AR-overexpressing SK-hep1 and HA22T cells and reversed the AR-modulated expression of c-Myc and p27. These results suggest that miR-135b-5p may suppress HCC proliferation via downregulation of AR-modulated HIF-2α, which in turn suppresses c-Myc expression, and increases expression of p27 in HCC cells. The ChIP assay and luciferase reporter assay further confirmed the direct molecular relationship by which AR transcriptionally modulates HIF-2α expression via its binding to the ARE located on the HIF-2α 5ʹ promoter in HCC. These experimental results provide a compelling narrative that miR-135b-5p suppresses androgen receptor-enhanced HCC in vitro via downregulating HIF-2α/c-Myc/p27 signals, which may help to better understand the HCC mechanism and develop more effective therapies to combat malignant HCC in the clinic.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This work was supported by NIH grants (CA155477 and CA156700), National Natural Science Foundation of China (81773105) and Natural Science Foundation of Anhui Province (1708085MH181). We thank Karen Wolf for help preparing the manuscript.
Disclosure
The author reports no conflicts of interest in this work. Shi-xiang Bao and Chun-hua Wang contributed equally.
References
1. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491. doi:10.1053/j.gastro.2018.08.065
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Feld JJ, Krassenburg LAP. What comes first: treatment of viral hepatitis or liver cancer? Dig Dis Sci. 2019;64(4):1041–1049. doi:10.1007/s10620-019-05518-5
4. ang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):
5. Ma WL, Lai HC, Yeh S, et al. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21(3):R165–R182.
6. Shi L, Yan P, Liang Y, et al. Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 2017;8(11):e3171.
7. Zhang H, Li -X-X, Yang Y, et al. Significance and mechanism of androgen receptor overexpression and androgen receptor/mechanistic target of rapamycin cross-talk in hepatocellular carcinoma. Hepatology. 2018;67(6):2271–2286. doi:10.1002/hep.29715
8. Yang WJ, Chang CJ, Yeh SH, et al. Hepatitis B virus X protein enhances the transcriptional activity of the androgen receptor through c-Src and glycogen synthase kinase-3beta kinase pathways. Hepatology. 2009;49(5):1515–1524. doi:10.1002/hep.22833
9. Xu J, Zheng L, Chen J, et al. Increasing AR by HIF-2α inhibitor (PT-2385) overcomes the side-effects of sorafenib by suppressing hepatocellular carcinoma invasion via alteration of pSTAT3, pAKT and pERK signals. Cell Death Dis. 2017;8(10):e3095. doi:10.1038/cddis.2017.411
10. Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57(2):840–847. doi:10.1002/hep.26095
11. Lagos-Quintana M, Rauhut R, Lendeckel W, et al. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–858. doi:10.1126/science.1064921
12. Lau NC, Lim LP, Weinstein EG, et al. An abundant class of tiny RNAs with probable regulatory roles in caenorhabditis elegans. Science. 2001;294(5543):858–862. doi:10.1126/science.1065062
13. Mollaei H, Safaralizadeh R, Rostami Z. MicroRNA replacement therapy in cancer. J Cell Physiol. 2019;234(8):12369–12384. doi:10.1002/jcp.28058
14. Xu JJ, Lin H, Li G, et al. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55–67. doi:10.1016/j.ebiom.2016.07.013
15. Ebron JS, Shukla GC. Molecular characterization of a novel androgen receptor transgene responsive to MicroRNA mediated post-transcriptional control exerted via 3ʹ-untranslated region. Prostate. 2016;76(9):834–844. doi:10.1002/pros.23174
16. Fletcher CE, Sulpice E, Combe S, et al. Androgen receptor-modulatory microRNAs provide insight into therapy resistance and therapeutic targets in advanced prostate cancer. Oncogene. 2019;38(28):5700–5724. doi:10.1038/s41388-019-0823-5
17. Liu C, Chen Z, Hu X, et al. MicroRNA-185 downregulates androgen receptor expression in the LNCaP prostate carcinoma cell line. Mol Med Rep. 2015;11(6):4625–4632. doi:10.3892/mmr.2015.3332
18. Wu H, Tao J, Li X, et al. MicroRNA-206 prevents the pathogenesis of hepatocellular carcinoma via modulating expression of cMET and Cdk6. Hepatology. 2017;66:1506–1511. doi:10.1002/hep.29374
19. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):
20. Tian F, Yu C, Wu M, et al. MicroRNA-191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 2019;52(5):e12635. doi:10.1111/cpr.12635
21. Zhai W, Sun Y, Jiang M, et al. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2α/C-MYC axis under hypoxia. Oncogene. 2016;35(37):4866–4880. doi:10.1038/onc.2016.19
22. Gallego F, Pisano MB, Torres C, et al. Molecular epidemiology of hepatitis B virus in Córdoba, Argentina. J Clin Virol. 2014;61(2):204–210. doi:10.1016/j.jcv.2014.06.030
23. White DL, Liu Y, Garcia J, et al. Sex hormone pathway gene polymorphisms are associated with risk of advanced hepatitis C-related liver disease in males. Int J Mol Epidemiol Genet. 2014;5(3):164–176.
24. Ma W-L, Lai H-C, Yeh S, et al. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21(3):165–182. doi:10.1530/ERC-13-0283
25. Och DC, Jang HS, O’Donnell EF, et al. Anti-androgen flutamide suppresses hepatocellular carcinoma cell proliferation via the aryl hydrocarbon receptor mediated induction of transforming growth factor-β1. Oncogene. 2015;34(50):
26. Dai B, Qu YY, Kong YY, et al. Kinetics of testosterone recovery in clinically localized prostate cancer patients treated with radical prostatectomy and subsequent short-term adjuvant androgen deprivation therapy. Asian J Androl. 2013;15(4):466–470. doi:10.1038/aja.2012.169
27. Ma WL, Hsu CL, Wu MH, et al. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135(3):
28. Wu MH, Ma WL, Hsu CL, et al. Androgen receptor promotes hepatitis B virus-induced hepatocarcinogenesis through modulation of hepatitis B virus RNA transcription. Sci Transl Med. 2010;2(32):32–35. doi:10.1126/scitranslmed.3001143
29. Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133(13):1487–1503. doi:10.1042/CS20190394
30. Lu XY, Chen D, Gu XY, et al. Predicting value of ALCAM as a target gene of microRNA-483-5p in patients with early recurrence in hepatocellular carcinoma. Front Pharmacol. 2018;8:973. doi:10.3389/fphar.2017.00973
31. Wei JX, Lv LH, Wan YL, et al. Vps4A functions as a tumor suppressor by regulating the secretion and uptake of exosomal microRNAs in human hepatoma cells. Hepatology. 2015;61(4):
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.