Back to Journals » OncoTargets and Therapy » Volume 12
miR-135a suppresses migration of gastric cancer cells by targeting TRAF5-mediated NF-κB activation
Authors Xie Y, Li F, Li Z, Shi Z
Received 6 October 2018
Accepted for publication 8 January 2019
Published 1 February 2019 Volume 2019:12 Pages 975—984
DOI https://doi.org/10.2147/OTT.S189976
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Yongzheng Xie,1,* Fangjun Li,2,* Zheng Li,1 Zhaohui Shi1
1Department of General Surgery, Henan University Huaihe Hospital, Kaifeng 475000, China; 2Department of Emergency, Henan University Huaihe Hospital, Kaifeng 475000, China
*These authors contributed equally to this work
Background: As crucial regulators and possible biomarkers for cancer development, miRNAs have attracted intensive attention during the last two decades. Among the known miRNAs, miR-135a has been indicated as a tumor suppressor in several cancer types, whereas its roles and mechanisms in gastric cancer (GC) remain largely unclear.
Materials and methods: Quantitative PCR (qPCR) was conducted to detect the expression of miR-135a in paired GC tissues as well as cell lines. The prognostic value was evaluated by Kaplan–Meier survival analysis. Wound healing and transwell assays were performed to determine the roles of miR-135a in GC cell migration. Dual-luciferase reporter assay, qPCR, and Western blot analysis were used to validate the targeting of TRAF5 and subsequent NF-κB pathway by miR-135a. Rescue experiments were done to explain the involvement of TRAF5 in mediating the anti-migration effect of miR-135a in GC cells. Finally, the expression of TRAF5 was examined in paired GC tissues.
Results: miR-135a was confirmed to be decreased in GC tissues and cell lines, and its lower expression predicted worse overall survival. Cellular experiments proved that miR-135a suppressed migration in GC cells. Through directly targeting TRAF5 and subsequently inhibiting NF-κB pathway, miR-135a might efficiently inhibit GC cell metastasis. Furthermore, we found that TRAF5 overexpression was negatively correlated with miR-135a expression in GC tissues.
Conclusion: Our study indicated that miR-135a serves a suppressing role in GC cell migration by targeting TRAF5 and the downstream NF-κB pathway.
Keywords: miR-135a, gastric cancer, TRAF5, NF-κB pathway, cell migration
Introduction
Gastric cancer (GC) is a common malignancy of the digestive system, and also represents the second frequent cause of cancer-related death worldwide.1,2 The incidence of GC has been increasing rapidly, with the highest incidence observed in northeast Asia.3 However, the mortality rate related to GC has seen a decline recently, largely because of the improvement of environment and diagnostic accuracy during the last few years.1,4 Nevertheless, it remains hard to be diagnosed at an early stage, which mainly limits the availability of therapy.5 Most of the GC patients are diagnosed at an advanced stage with metastasis to lymph nodes and distant organs, which leads to extremely poor clinical outcome of the medical treatment.6,7 Therefore, it is of great importance to reveal the molecular mechanisms of GC progression, especially the metastatic processes, to develop effective strategies against GC.
miRNAs are a group of conserved, short non-coding RNAs (19–25 nucleotides in length). Through binding to the 3′ UTR of the target mRNAs, they participate in gene expression regulation by blocking transcription or inducing transcriptional degradation.8–10 Numerous reports and reviews have summarized the roles of miRNAs in almost every aspect of cellular functions, including those in the physiological and pathological processes.11–14 The potential significance of miRNAs in cancer treatment is widely recognized.15–18
Depending on the target genes they regulate, miRNAs can act as oncogenes or tumor suppressors in GC.19,20 Intriguingly, the function of the same miRNA may be different in individual types of cancer, miR-135a is one such miRNA that belongs to this category. For instance, it is increased and promotes proliferation, migration, and invasion by targeting FOXO1 and TGFB1 in hepatocellular carcinoma,21,22 and it facilitates cell proliferation by targeting FOXO1 in malignant melanoma.23 However, it is decreased and inhibits migration and invasion by targeting KLF8 in lung cancer,24 it inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma,25 and it functions as a tumor suppressor in epithelial ovarian cancer.26 Notably, even in the same cancer, GC, scholars have respective opinions, and most studies identify miR-135a as a tumor suppressor.
In the present study, we focused on the effects of miR-135a on GC cell migration, which remains unclear until now. Our findings will provide new evidence for the role of miR-135a in GC and will certainly assist in the development of promising therapeutic targets for GC.
Materials and methods
Patients and tissues
Human GC tissues and paired adjacent normal tissues were acquired from 40 GC patients at Henan University Huaihe Hospital. After resection, all specimens were immediately snap-frozen in liquid nitrogen and stored at −80°C. Both GC tissues and paired adjacent normal tissues were histologically examined. This study was approved by the Ethics Committees of Henan University Huaihe Hospital and was conducted in accordance with the Declaration of Helsinki. All patients signed written informed consent.
Cell culture
Human GC cell lines (AGS, BGC-823, MKN-28, MKN-45, and SGC-7901) and human normal gastric mucous epithelium cells (GES-1) were purchased from the American Type Culture Collection (Manassas, VA, USA) or the Cell Bank of Chinese Academy of Science (Shanghai, China). All cells was cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), and supplemented with 10% FBS (Thermo Fisher Scientific), 100 U/mL penicillin, and 100 μg/mL streptomycin (P/S) at 37°C with 5% CO2 in a humidified incubator.
miRNA mimics, inhibitors, and transfections
miR-135a mimics, miR-135a inhibitor, miR-134-5p mimics, miR-134-5p inhibitor, or miRNA negative controls were purchased by GenePharma Company (Shanghai, China). Transfections were performed by using Lipofectamine 2000 reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions.
Wound healing assay
SGC-7901 cells transfected with miR-135a inhibitor and negative control were seeded in six-well plates and grown until 90% confluence. A 200 μL plastic pipette tip (Axygen, Union City, CA, USA) was used to scratch the monolayers. Then the wounded cells were washed twice with PBS and cultured in DMEM without serum for 24 hours. Images were taken to observe the wounds at the same fields under the microscope and the separation distance between wound sides was calculated. The result was shown as the percentage of wound closure, setting the initial scratch width as 100%.
Transwell assay
About 1×105 gastric cells were harvested, resuspended in serum-free medium, and further added to the top chamber of 24-well transwell chamber (Corning Incorporated, Corning, NY, USA) with a pore size of 8.0 μm. The lower chamber was filled with 500 μL of medium with 10% FBS. Twenty-four hours later, cells on the top chamber were removed by a cotton swab. Cells that migrated through the pores were fixed with paraformaldehyde for 20 minutes and stained with 0.5% trypan blue for 30 minutes at room temperature. The number of cells that migrated to the bottom surface was counted using a light microscope for at least five individual fields.
Real-time quantitative PCR (qPCR)
Total RNA was extracted from frozen tissues or GC cells using Trizol, and then cDNAs of miRNA were synthesized with Reverse Transcriptase M-MLV (RNase H-) (TaKaRa, Tokyo, Japan) by using specific primers (Ribobio, Guangzhou, China) as per the manufacturer’s instructions. The cDNAs of gene were synthesized with M-MLV Reverse Transcriptase (Promega Corporation, Fitchburg, WI, USA) as per the manufacturer’s instructions. Reaction mixture used for qPCR was Toyobo PCR Master Mix (Toyobo, Osaka, Japan). The relative expression levels of each miRNA or gene were calculated and normalized using the 2−ΔΔCt method relative to U6 or GAPDH.
Luciferase reporter assay
The wild-type 3′UTR of TRAF5 mRNA containing predicted miR-135a binding sites and mutant 3′UTR of TRAF5 were inserted into pmiRGLO vectors (Promega Corporation). BGC-823 or SGC-7901 cells were seeded in 24-well plates and cotransfected with wild-type or mutant 3′UTR of TRAF5 luciferase reporters and miRNA mimics or inhibitor using Lipofectamine 2000. After 48 hours, luciferase activity was detected using dual-luciferase reporter system (Promega Corporation) and normalized to Renilla activity. The coding sequence of TRAF5 mRNA was amplified by PCR using KOD-plus-Ver.2 kit (KOD-211; Toyobo) and cloned into pCMV-Tag2B expression vector. The NF-κB reporter plasmid was purchased from Beyotime Company (D2206 1 μg; Beijing, China). pRL Renilla Luciferase control reporter vectors were purchased from Promega Corporation.
Protein extraction and Western blot analysis
Total protein was extracted from cells by using RIPA buffer (Beyotime) supplemented with phenylmethylsulfonyl fluoride (Beyotime). BCA kit (Beyotime) was used to detect the concentration of protein in the supernatant. Protein expression was assessed by immunoblot analysis of 40 μg of cell lysate in the presence of antibodies to GAPDH, TRAF5, IκBα, p-p65 (Ser536), FAK, and JAK2 (Cell Signaling Technology, Danvers, MA, USA). Relative protein abundance of total protein was determined by normalizing to the endogenous control protein GAPDH.
Target prediction
Targets of miR-135a were searched on Targetscan Release 3.1 (http://www.targetscan.org/mamm_31/), and the results suggested TRAF5 is a potential target of miR-135a. To further confirm that TRAF5 is directly targeted by miR-135a, we obtained more information about the 3′UTR of TRAF5 mRNAs on Targetscan.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). Protein quantification was performed using ImageJ software (NIH, Bethesda, MD, USA). Statistical significance of difference between groups was determined by a two-tailed paired or grouped Student’s t-test. Kaplan–Meier analysis and log-rank test were used for survival analysis. The correlation between miR-135a and TRAF5 expressions was analyzed by Spearman’s correlation. All experiments were performed in triplicate and presented as mean ± SD. A P-value of <0.05 was considered as statistically significant.
Results
miR-135a is decreased in GC and associated with poor prognosis
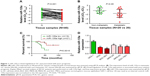
qPCR was first applied to examine the expression levels of miR-135a in 40 paired GC samples. Our results confirmed its downregulation in GC tissues compared with paired adjacent normal tissues (Figure 1A). Among these 40 GC samples, metastatic GC tissues (N=20) contained relatively lower levels of miR-135a in comparison with those of non-metastatic GC tissues (N=20; Figure 1B). These findings suggested a potential association of miR-135a with GC progression and metastasis. In addition, patients with lower miR-135a levels exhibited shorter survival time, whereas those with higher miR-135a levels exhibited longer survival time (Figure 1C). This finding highly indicated that miR-135a might be taken as a novel prognostic predicator for GC patients. Our following examination of its expression in GC cell lines further demonstrated that miR-135a was decreased in GC cell lines (AGS, BGC-823, MKN-28, MKN-45, and SGC-7901) compared with that in human normal gastric mucous epithelium cells (GES-1) (Figure 1D). Therefore, these results indicated that miR-135a is indeed decreased in GC tissues and cell lines, and that it might play an important role in GC progression and metastasis.
miR-135a inhibits GC cell migration in vitro
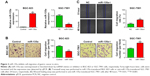
To investigate the effect of miR-135a on GC cell migration, we chose BGC-823 and SGC-7901 cell lines to do gain- and loss-of-function assays, respectively. We transfected BGC-823 cells with miR-135a mimics or negative control mimics (control), and transfected SGC-7901 cells with miR-135a inhibitor or negative control inhibitor (NC). The transfection efficiency was confirmed by qPCR (Figure 2A). Cell migration abilities were assessed by transwell assay and wound healing assay. We found that overexpressing miR-135a in BGC-823 cells reduced the migratory capacity, while inhibiting endogenous miR-135a in SGC-7901 cells elevated the migratory capacity (Figure 2B and C). Therefore, we concluded from the above-mentioned results that miR-135a inhibits GC cell migration in vitro.
TRAF5 is a direct target of miR-135a in GC cells
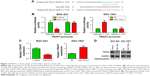
To understand the mechanism of action of miR-135a in GC cell migration, we conducted bioinformatics analysis of the targets of miR-135a based on the database TargetScan Release 3.1. Among these predicted targets, we noticed that TRAF5 was the putative target of miR-135a (Figure 3A). The 3′ UTR of TRAF5 mRNA contains a potential binding site for miR-135a. We then constructed two luciferase reporters containing wild-type or mutant 3′ UTR of TRAF5 mRNA (only the region spanning the potential binding site of miR-135a; Figure 3A). The dual-luciferase reporter assay revealed that co-transfection of miR-135a mimics and wild-type 3′ UTR of TRAF5 promoter significantly reduced the luciferase activity. However, co-transfection of miR-135a mimics and mutant 3′ UTR of TRAF5 promoter retained the similar luciferase activity as the control. Meanwhile, co-transfection of miR-135a inhibitor with wild-type 3′ UTR of TRAF5 promoter significantly induced the luciferase activity (Figure 3B). qPCR and Western blot analysis validated the finding that overexpressing miR-135a inhibited TRAF5 mRNA and protein expression in BGC-823 cells, while inhibiting endogenous miR-135a elevated TRAF5 mRNA and protein expression in SGC-7901 cells (Figure 3C and D). In order to be more rigorous, irrelevant miRNA (miR-134-5p) was served as another NC. The results showed that miR-134-5p did not alter the protein level of TRAF5 in both BGC-823 and SGC-7901 cells (Figure S1). Collectively, our results indicated that miR-135a negatively regulates the expression of TRAF5 in GC cells by base pairing to the 3′ UTR of TRAF5 mRNA.
miR-135a inhibits the activity of NF-κB signaling pathway
Considering the connection of TRAFs with classical NF-κB pathway,27 we next wondered whether NF-κB pathway is involved in the anti-migration effects of miR-135a in GC cells. Western blot analysis showed that overexpressing miR-135a in BGC-823 cells inhibited the expression of phospho-p65 and increased the expression of IκBα (Figure 4A). Luciferase reporter assay also verified that miR-135a suppressed the NF-κB reporter activity (Figure 4B). qPCR detection of the related targets of NF-κB signaling in the aspect of metastasis controlling confirmed that miR-135a inhibited the mRNA levels of MMP2, MMP9, ICAM-1, and VCAM-1 (Figure 4C). Furthermore, inhibiting endogenous miR-135a in SGC-7901 cells had the opposite effects as revealed by Western blot, luciferase reporter, and qPCR assays mentioned earlier (Figure 4D–F). Hence, we proposed that miR-135a inhibits NF-κB pathway through directly targeting TRAF5, since TRAF5 could activate this pathway.
Restoration of TRAF5 rescued the anti-migration effects of miR-135a in GC
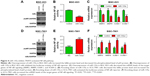
To understand the bridge role of TRAF5 in miR-135a-mediating anti-migration and NF-κB signaling pathway inhibition, we introduced exogenous TRAF5 into miR-135a overexpressed BGC-823 cells by co-transfection of TRAF5 plasmid and miR-135a mimics (Figure 5A). Western blot analysis showed that TRAF5 efficiently rescued the decrease of p-p65 and increase of IκBα (Figure 5A). Luciferase reporter and qPCR assays also confirmed the rescue of NF-κB signaling pathway by TRAF5 (Figure 5B and C). More importantly, restoration of TRAF5 in miR-135a overexpressed BGC-823 cells significantly reversed the anti-migration effects caused by miR-135a (Figure 5D). Collectively, our results indicated that TRAF5 attenuates miR-135a-induced suppression of GC cell migration through NF-κB signaling pathway. In other words, miR-135a inhibits GC cell migration by targeting TRAF5 and subsequently blocking NF-κB pathway.
TRAF5 is overexpressed and negatively correlated with miR-135a expression
Finally, we examined the expression levels of TRAF5 in the above-mentioned 40 paired GC samples. qPCR results showed that TRAF5 mRNA was upregulated in GC tissues compared with paired adjacent normal tissues (Figure 6A). We also observed that metastatic GC tissues (N=20) contained relatively higher levels of TRAF5 in comparison with those of non-metastatic GC tissues (N=20; Figure 6B). Importantly, TRAF5 mRNA levels negatively correlated with the expression of miR-135a, highlighting the targeting of TRAF5 by miR-135a in GC samples and cell lines (Figure 6C and D). Therefore, these results indicated that TRAF5 itself is overexpressed in GC samples, associated with metastasis status, and might participate in GC progression.
Discussion
Metastasis is the major challenge limiting the therapeutic effect of human cancers and also the overwhelming cause of cancer mortality.6,7,28 Although this point is well recognized by scholars for decades, a comprehensive picture of cellular and molecular determinants governing cancer metastasis remains faint. Containing a series of cellular behaviors including epithelial–mesenchymal transition, migration, and invasion, metastasis is under strict control by coding and non-coding RNAs.28 Previous reports have demonstrated that miRNAs are essential for cancer metastasis by targeting related mRNA targets.29
In the present study, miR-135a was found to be decreased in GC tissues, especially in those from metastatic GC samples, and its lower expression was associated with worse prognosis in GC patients. Following cellular experiments proved that miR-135a could suppress cell migration. These findings enrich the roles of miR-135a in GC cells, and indicate that miR-135a may serve as a biomarker or therapeutic target for GC. Indeed, the correlation of miR-135a with GC metastasis was firstly implied by Liu et al, since they showed that the expression of miR-135a was lower in GC tissues with lymph node metastasis.30 This connection was reinforced by a later study, in which Cheng et al found that decreased miR-135a levels in GC is associated with TNM stage and poor survival. They also found that miR-135a significantly inhibits cell growth, migration, invasion, and angiogenesis by targeting focal adhesion kinase pathway.31 Moreover, in early reports, Wu et al indicated that miR-135a targets JAK2 and inhibits GC cell proliferation.32 We also detected these two targets, but did not found any changes in our system (Figure S2). In 2016, Yan et al showed the tumor promoting function of miR-135a in drug resistance. They reported that miR-135a could promote GC progression and resistance to oxaliplatin by inhibiting E2F1 expression and the Sp1/DAPK2 signaling pathway.33 We suppose this opposite functions of miR-135a may be caused by the different targets and different pathways involved. Overall, our results presented here strengthened their conclusion that miR-135a acts as an anti-metastasis factor in GC.
Recent work has established that TRAF5 is targeted by several miRNAs and involved in cancer cell proliferation, apoptosis, and metastasis.34,35 In agreement with these findings, we found that TRAF5 is targeted by miR-135a based on the following observations: 1) dual-luciferase reporter assay, qPCR assay, and Western blot analysis clearly verified that miR-135a targets TRAF5 in GC cells; 2) qPCR validated that TRAF5 is overexpressed in GC tissues and negatively correlated with miR-135a expression. This targeting provides a novel explanation for the anticancer roles of miR-135a.
The transcription factor NF-κB has attracted extensive attention in cancer research. Its excessive activation is frequently observed occurred in human cancers, and its family members as well as the downstream target genes are linked to various types of malignancy phenotypes, including proliferation, migration, invasion, and therapeutic resistance.36 TRAFs are linked with NF-κB pathway activation,27 and Tao et al had proved in glioma cells that TRAF5 is required for the activation of NF-κB.37 We herein confirmed in GC cells that miR-135a inhibits NF-κB signaling pathway. Combined with the finding that miR-135a directly targets TRAF5, we can safely conclude that miR-135a inhibits NF-κB pathway through directly targeting TRAF5, since TRAF5 could activate this pathway. Restoration of TRAF5 in miR-135a overexpressed GC cells rescued both NF-κB signaling and cell migration, further highlighting that miR-135a inhibits GC cell migration by targeting TRAF5 and subsequently blocking NF-κB pathway.
In summary, our study confirmed that miR-135a is decreased in GC and exerts anti-migration effects by targeting TRAF5 and downstream NF-κB signaling pathway. Both miR-135a and TRAF5 might be considered as biomarkers and/or therapeutic targets for GC.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. | ||
Shah MA. Update on metastatic gastric and esophageal cancers. J Clin Oncol. 2015;33(16):1760–1769. | ||
Hartgrink HH, Jansen EPM, van Grieken NCT, van de Velde CJH. Gastric cancer. Lancet. 2009;374(9688):477–490. | ||
Purushotham AD, Lewison G, Sullivan R. The state of research and development in global cancer surgery. Ann Surg. 2012;255(3):427–432. | ||
Baastrup R, Sørensen M, Hansen J, Hansen RD, Würtzen H, Winther JF. Social inequality and incidence of and survival from cancers of the oesophagus, stomach and pancreas in a population-based study in Denmark, 1994–2003. Eur J Cancer. 2008;44(14):1962–1977. | ||
Piazuelo MB, Correa P. Gastric cáncer: overview. Colomb Med (Cali). 2013;44(3):192–201. | ||
Jiang Y, Ajani JA. Multidisciplinary management of gastric cancer. Curr Opin Gastroenterol. 2010;26(6):640–646. | ||
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. | ||
O’Brien J, Hayder H, Zayed Y, Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne). 2018;9:402. | ||
Liu W, Ma R, Yuan Y. Post-transcriptional regulation of genes related to biological behaviors of gastric cancer by long noncoding RNAs and microRNAs. J Cancer. 2017;8(19):4141–4154. | ||
Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. | ||
Cha W, Fan R, Miao Y, et al. MicroRNAs as novel endogenous targets for regulation and therapeutic treatments. Medchemcomm. 2018;9(3):396–408. | ||
Piotto C, Julier Z, Martino MM. Immune regulation of tissue repair and regeneration via miRNAs–new therapeutic target. Front Bioeng Biotechnol. 2018;6:98. | ||
Haider MT, Taipaleenmäki H. Targeting the metastatic bone microenvironment by microRNAs. Front Endocrinol (Lausanne). 2018;9:202. | ||
Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. Epub 2018 Aug 17. | ||
Zhang Y, Xu B, Zhang XP. Effects of miRNAs on functions of breast cancer stem cells and treatment of breast cancer. Onco Targets Ther. 2018;11:4263–4270. | ||
To KK, Tong CW, Wu M, Cho WC. microRNAs in the prognosis and therapy of colorectal cancer: from bench to bedside. World J Gastroenterol. 2018;24(27):2949–2973. | ||
Guo S, Fesler A, Wang H, Ju J. microRNA based prognostic biomarkers in pancreatic cancer. Biomark Res. 2018;6(1):18. | ||
Link A, Kupcinskas J. MicroRNAs as non-invasive diagnostic biomarkers for gastric cancer: current insights and future perspectives. World J Gastroenterol. 2018;24(30):3313–3329. | ||
Yuan HL, Wang T, Zhang KH. MicroRNAs as potential biomarkers for diagnosis, therapy and prognosis of gastric cancer. Onco Targets Ther. 2018;11:3891–3900. | ||
Zeng YB, Liang XH, Zhang GX, et al. miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 2016;16(1):63. | ||
Yao S, Tian C, Ding Y, et al. Down-regulation of Krüppel-like factor-4 by microRNA-135a-5p promotes proliferation and metastasis in hepatocellular carcinoma by transforming growth factor-β1. Oncotarget. 2016;7(27):42566–42578. | ||
Ren JW, Li ZJ, Tu C. miR-135 post-transcriptionally regulates FoxO1 expression and promotes cell proliferation in human malignant melanoma cells. Int J Clin Exp Pathol. 2015;8(6):6356–6366. | ||
Shi H, Ji Y, Zhang D, Liu Y, Fang P. miR-135a inhibits migration and invasion and regulates EMT-related marker genes by targeting KLF8 in lung cancer cells. Biochem Biophys Res Commun. 2015;465(1):125–130. | ||
Dang Z, Xu WH, Lu P, et al. MicroRNA-135a inhibits cell proliferation by targeting Bmi1 in pancreatic ductal adenocarcinoma. Int J Biol Sci. 2014;10(7):733–745. | ||
Tang W, Jiang Y, Mu X, Xu L, Cheng W, Wang X. miR-135a functions as a tumor suppressor in epithelial ovarian cancer and regulates HOXA10 expression. Cell Signal. 2014;26(7):1420–1426. | ||
Sakurai H, Suzuki S, Kawasaki N, et al. Tumor necrosis factor-alpha-induced IKK phosphorylation of NF-kappaB p65 on serine 536 is mediated through the TRAF2, TRAF5, and TAK1 signaling pathway. J Biol Chem. 2003;278(38):36916–36923. | ||
Seyfried TN, Huysentruyt LC. On the origin of cancer metastasis. Crit Rev Oncog. 2013;18(1–2):43–73. | ||
di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol Mech Dis. 2014;9(1):287–314. | ||
Liu L, Ye JX, Qin YZ, Chen QH, Ge LY. Evaluation of miR-29c, miR-124, miR-135a and miR-148a in predicting lymph node metastasis and tumor stage of gastric cancer. Int J Clin Exp Med. 2015;8(12):22227–22236. | ||
Cheng Z, Liu F, Zhang H, et al. miR-135a inhibits tumor metastasis and angiogenesis by targeting FAK pathway. Oncotarget. 2017;8(19):31153–31168. | ||
Wu H, Huang M, Cao P, Wang T, Shu Y, Liu P. miR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol Ther. 2012;13(5):281–288. | ||
Yan LH, Chen ZN, Li-Li, et al. miR-135a promotes gastric cancer progression and resistance to oxaliplatin. Oncotarget. 2016;7(43):70699–70714. | ||
Li M, Long C, Yang G, Luo Y, Du H. miR-26b inhibits melanoma cell proliferation and enhances apoptosis by suppressing TRAF5-mediated MAPK activation. Biochem Biophys Res Commun. 2016;471(3):361–367. | ||
Chen Z, Zhao L, Zhao F, Yang G, Wang J. MicroRNA-26b regulates cancer proliferation migration and cell cycle transition by suppressing TRAF5 in esophageal squamous cell carcinoma. Am J Transl Res. 2016;8(5):1957–1970. | ||
Li F, Zhang J, Arfuso F, et al. NF-κB in cancer therapy. Arch Toxicol. 2015;89(5):711–731. | ||
Tao T, Cheng C, Ji Y, et al. Numbl inhibits glioma cell migration and invasion by suppressing TRAF5-mediated NF-κB activation. Mol Biol Cell. 2012;23(14):2635–2644. |
Supplementary materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.