Back to Journals » OncoTargets and Therapy » Volume 13
miR-133b Suppresses Invasion and Migration of Gastric Cancer Cells via the COL1A1/TGF-β Axis
Authors Guo Y, Lu G, Mao H, Zhou S, Tong X, Wu J, Sun Q, Xu H, Fang F
Received 13 February 2020
Accepted for publication 17 July 2020
Published 12 August 2020 Volume 2020:13 Pages 7985—7995
DOI https://doi.org/10.2147/OTT.S249667
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Yuan Guo, Guochun Lu, Huahui Mao, Shengkun Zhou, Xiangmei Tong, Junfei Wu, Qiang Sun, Hui Xu, Fu Fang
Department of General Surgery, First People’s Hospital of Tonglu, Hangzhou 311500, People’s Republic of China
Correspondence: Fu Fang
Department of General Surgery, First People’s Hospital of Tonglu, No. 338 Xuesheng Road, Hangzhou 311500, People’s Republic of China
Tel +86-15168291988
Email [email protected]
Objective: The study aimed to explore the mechanism of miR-133b regulating the invasion and migration of gastric cancer (GC) cells via the COL1A1/TGF-β axis.
Methods: The miRNA expression profiles of GC downloaded from TCGA database were subjected to differential analysis to determine the target miRNA of interest, and the target genes of the miRNA were predicted by bioinformatics. GSEA was used for gene enrichment analysis. qRT-PCR was carried out to detect gene expression in GC cells. The effect of miR-133b on GC cells was examined by CCK-8, wound healing and Transwell assays. Western blot was conducted to assess the protein expression of EMT-related proteins. The binding relationship between genes was verified by dual-luciferase reporter gene assay.
Results: The expression of miR-133b was markedly downregulated in GC tissue, while that of COL1A1 was upregulated. Overexpression of miR-133b decreased the migration and invasion of GC cells, and the EMT process was inhibited as well, while inverse results were observed when miR-133b was silenced. COL1A1 was a target gene of miR-133b and its overexpression had a significant impact on the prognosis of patients. GSEA pathway enrichment results showed that COL1A1 was markedly enriched in the TGF-β signaling pathway. In addition, COL1A1 overexpression induced the activation of the TGF-β signaling pathway to promote proliferation and migration of GC cells, whereas miR-133b overexpression suppressed the signaling pathway. Thus, overexpression of miR-133b and COL1A1 simultaneously would reverse the inhibitory effect of miR-133b on cell invasion and migration.
Conclusion: In this study, miR-133b was found to inhibit the invasion and migration of GC cells via the COL1A1/TGF-β axis, which provides a new research direction for the diagnosis and targeted therapy of GC.
Keywords: gastric cancer, miR-133b, COL1A1, TGF-β signaling pathway, invasion and migration
Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed cancer worldwide. According to statistics, there were more than 1 million new global GC cases in 2018, with an estimate of 783 thousand deaths.1 Genetic mutations and environmental factors such as helicobacter pylori infection have been recognized as the main cause leading to GC occurrence, while smoking and alcohol consumption have also been identified to be implicated in.1,2 With the development of medical treatment in recent years, chemotherapy can alleviate the symptoms of some GC patients, which improves the survival and life quality of sufferers. However, about 60% of patients are diagnosed with advanced GC, with limited treatment options and poor prognosis. At primary presentation, stage IV metastatic GC is often diagnosed, which causes poor outcomes, and it is incurable with a very poor prognosis (5-year survival rate of ~ 4%).3 Therefore, it is very important to study the mechanism that regulates the progression of GC and to explore new diagnostic and therapeutic targets.
miRNAs are small non-coding RNAs that regulate gene expression by targeting the 3ʹ-UTR of specific mRNA.4 In recent years, increasing studies have shown that the dysregulation of miRNA expression plays an important role in the development of multiple cancers, including GC.5–7 miR-133b has been reported to be downregulated and can be used as a tumor suppressor gene in various cancers, including breast cancer,8 bladder cancer,9 colorectal cancer10 and esophageal squamous cell cancer.11 However, studies on GC have reported that knock down the expression of miR-133b promotes the proliferation of cancer cells,9 and circulating miR-133b can be introduced as a potential diagnostic marker for GC.12 Nevertheless, the mechanism of miR-133b regulating GC metastasis has been less studied. So, we conducted the research to further explore the mechanism.
Collagen type I α 1 (COL1A1) is a member of collagen family, and evidence has proved that the collagen family members promote the progression of multiple cancers.13 In addition, the abnormal expression of COL1A1 has also been confirmed to be associated with the occurrence of some cancers.14–16 COL1A1 is a reliable biomarker and a potential therapeutic target for hepatocellular carcinogenesis and metastasis.17 However, the role and the molecular regulation mechanism of COL1A1 in GC development remain unclear. Moreover, no research has found the regulatory relationship between COL1A1 and miR-133b. TGF-β signaling has been demonstrated to regulate several biological processes, whose abnormal activation contributes to tumor development.18 It is reported that there is a positive regulatory relationship between COL1A1 and TGF-β signaling.19–21 However, there is no similar finding in GC.
This study is designed to explore the relationship between miR-133b and GC, reveal the molecular mechanism of miR-133b targeted regulating COL1A1, and to provide a new research direction for targeted therapy of GC.
Materials and Methods
Bioinformatics Analysis
The miRNA (normal: n=45, tumor: n=444) and mRNA (normal: n=32, tumor: n=373) expression profiles of TCGA-STAD were downloaded from TCGA database (https://portal.gdc.cancer.gov/), and differential analysis was conducted to identify the target miRNA by using edgeR package with |logFC|>2 and padj<0.05 as the threshold. Differentially expressed mRNAs (DEmRNAs) and the target genes of the miRNA predicted by targetScan, miRDB, and mirDIP databases were intersected to obtain the mRNAs bearing targeted binding sites with the miRNA. Survival analysis of target genes was conducted based on clinical information, and pathway enrichment analysis was conducted by GSEA software to study the role of the target miRNA and mRNA in GC.
Cell Culture
Human normal gastric epithelial cell line GES-1 (BNCC337970) and 5 GC cell lines AGS (BNCC338141), SGC-7901 (BNCC100674), MGC-803 (BNCC100665), HGC-27 (BNCC100716) and BGC-823 (BNCC337689) (BeNa Culture Collection, China) were cultured in Dulbecco’s Modified Eagle’s medium (DMEM; Thermo Scientific HyClone, China) containing 5% fetal bovine serum (FBS) and placed in a humidified incubator (TealMo, USA) with 5% CO2 at 37 °C.
Cell Transfection
NC mimic, miR-133b mimic, NC inhibitor and miR-133b inhibitor were obtained from GenePharma (Shanghai, China). The sequences of oe-NC (negative control vector), oe-COL1A1(COL1A1 overexpression vector) were synthesized by Sangon Biotech (Shanghai) Co., Ltd. The recombinant plasmid pEGFP1-COL1A1 was constructed using the pEGFP1 overexpression vector. Cells in 3×105 cells/well were seeded into a six-well plate and transfected when the growth density reached 50% according to the instructions of lipofectamin2000 (11,668–019, Invitrogen, California, USA). Four micrograms of recombinant plasmids and 10 μL Lipofectamin2000 were diluted by 250 μL Opti-MEM medium (51,985,042, Gibco, Gaithersburg, MD, USA) without serum, respectively. After keeping at room temperature for 5 min, the two liquids were mixed together uniformly and added dropwise to the cell culture wells 20 min later. After shaking, cells were cultured in a cell incubator with 5% CO2 at 37 °C. Six h later, the medium was replaced and cells were harvested 48 h after transfection.
qRT-PCR
Total RNA in cells was extracted by TRIzol Reagent (Invitrogen), and 2 μg total RNA was inverted into cDNA by Superscript II reverse transcriptase (Invitrogen). qRT-PCR was performed using the miScript SYBR Green PCR Kit (Qiagen, Hilden, Germany) on the Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, USA) to detect the expression of miR-133b and COL1A1. All primers shown in Table 1 were purchased from Sangon Biotech (Shanghai) Co., Ltd. miRNA and mRNA used U6 and GAPDH as internal regulators, respectively. The differences in the relative expressions of the target mRNA and miRNA in the control and experimental groups were compared by 2−ΔΔCt method.
 |
Table 1 Primer Sequences |
Western Blot (WB)
Forty-eight hours after transfection, cells in each group were washed 3 times with cold PBS and lysed on ice for 10 min with whole cell lysate. According to the manufacturer’s instructions, nuclear extracts were prepared using the Nuclear Extraction Kit (Active Motif) prior to the detection of nucleoproteins. Protein concentration was determined using the BCA kit (Thermo Fisher Scientific, USA). A total of 30 μg proteins were loaded onto SDS-PAGE electrophoresis. Subsequently, the proteins were transferred onto PVDF membrane (Amersham, USA), which was sequentially blocked with 5% bovine serum albumin (BSA; AMRESCO, Solon, OH, USA) at room temperature for 1 h. After discarding the blocking buffer, the membrane was incubated with primary antibodies including COL1A1 rabbit polyclonal antibody (abcam, Cambridge, UK), N-cadherin rabbit polyclonal antibody (abcam), E-cadherin rabbit polyclonal antibody (abcam), TGFβR1 rabbit polyclonal antibody (abcam), TGFβR2 rabbit polyclonal antibody (abcam), SMAD3 (abcam), p-SMAD3 rabbit monoclonal antibody (Cell Signaling Technology), GADPH rabbit polyclonal antibody (abcam) and p84 (abcam) at 4 °C overnight. p84 was used as a reference for nuclear extractions, and GAPDH was the reference for other proteins. The membrane was washed with PBST for 3 times for 10 min of each time. Then, goat anti-rabbit IgG H&L (HRP) (ab6721, 1:3000, abcam) was added to the membrane for incubation. Later, the membrane was washed with PBST following the above steps.
Wound Healing Assay
When cells grew at 70–80% in confluence in the culture plate, a scratch was made through the center of each well using a 200 μL pipette tip. The dislodged cells were removed after the plate was washed twice with PBS. Cells were then cultured with fresh medium and photographed under a microscope after another 24 h of growth to calculate the migration rate.
Transwell Invasion Assay
A 24-well Transwell chamber (8 μm pore size, BD Biosciences) was used for Transwell invasion assay. The upper chamber coated with Matrigel (Corning, Corning, NY) was added with approximately 2×104 cells and the lower chamber was filled with DMEM containing 10% FBS. After incubation at 37 °C for 24 h, the cells on the upper surface of the membrane were removed with a cotton swab, and the cells in the lower chamber were stained with crystal violet. Then, four fields were randomly selected under a microscope to calculate the number of cells that successfully invaded the matrix matrigel.
Dual-Luciferase Reporter Gene Assay
The corresponding mutant sequence was created through substituted mutating the regions of the miR-133b binding sites. The mutant-type (MUT) or wild-type (WT) 3ʹ-UTR of COL1A1 was amplified and cloned into the downstream of pmirGLO (Promega, WI, USA) Luciferase reporter vector to construct the COL1A1-WT and COL1A1-MUT groups. Renilla luciferase expression vector pRL-TK (TaKaRa, Dalian, China) was used as an internal reference. Afterwards, the target plasmid was co-transfected with the luciferase reporter vectors into the HEK-293T cells. The relative luciferase activity was calculated using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s procedures.
Statistical Analysis
All data were processed using SPSS 21.0 statistical software (SPSS; Inc, Chicago, IL, USA). The measurement data were expressed in the form of mean ± standard deviation. Student’s t-test was used to analyze the differences between the two groups. Kaplan–Meier method was used to calculate the overall survival, and Log-rank test was used to analyze the difference in patient survival. P<0.05 indicated that the difference was statistically significant, and P<0.01 indicated that the difference was highly significant. All experiments were repeated three times.
Results
miR-133b is Downregulated While COL1A1 is Upregulated in GC Tissue and Cells
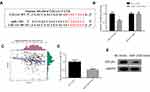
A total of 103 miRNAs (DEmiRNAs) and 1678 DEmRNAs were obtained by differential analysis (Figure 1A and B). Through analyzing the expression of miR-133b in TCGA database, we found it was downregulated in tumor tissue (Figure 1C, P<0.05). miR-133b has also been proved to express differentially in multiple cancer tissues, and get involved in the regulation of tumor cell invasion and migration.8,11 Therefore, miR-133b was selected as the target miRNA. The target genes of miR-133b were predicted and intersected with 901 upregulated DEmRNAs to obtain 4 DEmRNAs (COL1A1, FOXL2, HOXA9, POU4F1) with binding sites of miR-133b (Figure 1D). Through bioinformatics analysis, we found that compared with normal tissue, COL1A1 was significantly upregulated in tumor tissue (Figure 1E, P<0.05), which was related to poor prognosis of patients by survival analysis (Figure 1F). qRT-PCR was used to further detect the expression of miR-133b and COL1A1 in normal gastric cell line GES-1 and GC cell lines, and the results confirmed that miR-133b was significantly downregulated while COL1A1 was upregulated in GC cell lines. With the lowest miR-133b expression and the highest COL1A1 expression (Figure 1G and H, P<0.01), the HGC-27 cell line was selected for the subsequent experiments.
miR-133b Suppresses Invasion and Migration of GC Cells
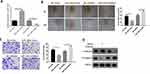
miR-133b mimic and inhibitor were transfected into HGC-27 cells to detect their effects on cell invasion and migration. Firstly, we detected the expression of miR-133b in cells of each group by qRT-PCR after transfection and the results displayed that the expression of miR-133b was remarkably increased in miR-133b mimic group, while that in the inhibitor group was suppressed (Figure 2A). Subsequently, cell migration and invasion were examined by the wound healing (Figure 2B) and the Transwell assays (Figure 2C). After transfection with miR-133b mimic, cell migration and invasion abilities were decreased significantly, while reverse results were observed after cells were transfected with miR-133b inhibitor. The results were also verified by WB from protein level. Additionally, the E-cadherin protein level was increased in the miR-133b mimic group, while N-cadherin was decreased, suggesting that the epithelial–mesenchymal transition (EMT) process in cancer cells was inhibited. While the expression of these two EMT-related proteins in the inhibitor group showed opposite trends (Figure 2D), the above results demonstrated that miR-133b played a role in inhibiting migration and invasion in GC cells.
miR-133b Targeted Downregulates COL1A1 Expression
Previously, we found that miR-133b may target the downstream gene COL1A1. To further verify the targeted binding relationship between miR-133b and COL1A1, we predicted the binding site sequences through starBase database (Figure 3A) and verified the relationship through dual-luciferase assay (Figure 3B). The results observed that miR-133b was able to bind to the 3ʹ-UTR of COL1A1-WT, and the luciferase activity of cells with miR-133b mimic and COL1A1-WT vector co-transfection group was significantly inhibited, but no change was observed after cells were transfected with COL1A1-MUT. We further analyzed the expression correlation between miR-133b and COL1A1, and found that miR-133b was remarkably negatively correlated with COL1A1 (Figure 3C), which was further verified by qRT-PCR and WB (Figure 3D and E). These results exhibited that miR-133b inhibited COL1A1 expression in GC cells.
COL1A1 Promotes Invasion and Migration of GC Cells via Activating the TGF-β Signaling Pathway
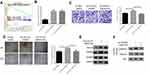
The results of GSEA pathway enrichment analysis showed that COL1A1 was significantly enriched in the TGF-β signaling pathway (Figure 4A), and it has been reported that COL1A1 is involved in regulating the TGFβ pathway in bladder cancer cells.21 Therefore, we speculated that COL1A1 could activate the TGF-β pathway to promote the invasion and migration of GC cells. GC cells were divided tinto three groups: oe-NC+DMSO, oe-COL1A1+DMSO, and oe-COL1A1+SB431542 (SB431542, TGF-β/Smad inhibitor, Adooq Bioscience, A10826-10) to detect cell migration, invasion, and TGF-β signaling pathway-related protein expression. Firstly, we detected the expression of COL1A1 in each group. The results showed that compared with the oe-NC+DMSO group, the mRNA and protein expression of COL1A1 in the oe-COL1A1+DMSO group were increased, while the addition of SB431542 did not affect COL1A1 expression (Figure 4B and E). Then, we detected the invasion and migration abilities of cells through Transwell (Figure 4C) and wound healing assays (Figure 4D). The results indicated that overexpression of COL1A1 significantly improved the migration and invasion abilities of GC cells, while the addition of SB431542 reversed the promoting effect of COL1A1. We further examined the expression of TGFβR1, TGFβR2 (Figure 4E) and the nuclear proteins pSMAD3 and SMAD3 (Figure 4F) associated with the TGF-β signaling pathway. It was found that compared with the oe-NC+DMSO group, the expression of TGFβR1, TGFβR2 and pSMAD3 in the oe-COL1A1+DMSO group were significantly increased. Notably, the expression of these proteins in the oe-COL1A1+SB431542 group with the addition of SB431542 was significantly decreased. The above results indicated that COL1A1 promoted the migration and invasion of GC cells by activating the TGF-β pathway.
miR-133b Suppresses Invasion and Migration of GC Cells via the COL1A1/TGF-β Axis
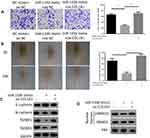
In order to verify the mechanism of miR-133b regulating the COL1A1/TGF-β axis to affect the occurrence and development of GC, we conducted rescue experiments. At first, we detected the invasion and migration abilities in three groups (Figure 5A and B). The results revealed that compared with NC mimic+oe-NC group, the migration and invasion abilities of cells in miR-133b mimic+oe-NC group were significantly reduced, while the abilities were remarkably enhanced in miR-133b mimic+oe-COL1A1 group. We further tested the expression of EMT-related proteins, E-cadherin and N-cadherin, as well as TGF-β signaling pathway-related proteins (Figure 5C and D). The results displayed that overexpression of miR-133b decreased the expression of N-cadherin, TGFβR1, TGFβR2, and pSMAD3, while overexpressing miR-133b and COL1A1 simultaneously reversed the expression of these proteins, which were similar to those in NC mimic+oe-NC group. The above results indicated that miR-133b inhibited the invasion and migration of GC cells via the COL1A1/TGF-β axis.
Discussion
A growing number of studies have suggested that miRNA dysregulation can regulate tumor proliferation and metastasis. miR-133b has been identified to act as a tumor suppressor gene in various cancers and is closely related to suppressed tumor metastasis. For example, miR-133b is significantly downregulated in esophageal squamous cell carcinoma (ESCC), and miR-133b/EGFR axis regulates cell metastasis by suppressing anoikis resistance and anchorage-independent growth.22 Huang et al discovered that the overexpression of REST inhibits miR-133 transcription in prostate cancer tissue, thereby contributing to enhanced invasion and migration abilities in vitro and bone metastasis ability in vivo via activating the TGF-β signaling.23 In this study, we found that miR-133b expression was dramatically lower in tumor tissue than that in normal tissue by bioinformatics. Furthermore, we observed that miR-133b expression in GC cells was remarkably lower than that in gastric epithelial cells by in vitro experiments. Overexpression of miR-133b suppressed invasion and migration of GC cells and the occurrence of EMT was also inhibited, while opposite results were observed when miR-133b was silenced. Our study indicates that the development of GC is most likely related to the dysregulation of miR-133b expression, which can be supported by the research results in other cancers.
Mining the direct targets of miRNAs is essential for studying cancer development, and helps to clarify the regulatory pathways and mechanisms of miRNAs. Here, we found that COL1A1 may be the direct target gene of miR-133b. In the meantime, COL1A1 was found to be upregulated in GC tissue and its expression was negatively related to the prognosis of patients by bioinformatics. Next, we verified the targeted binding relationship between COL1A1 and miR-133b through dual-luciferase assay. COL1A1 as a member of the collagen family is differentially expressed in multiple cancers.24–26 A study demonstrated that the expression of COL1A1 is significantly increased in cervical cancer tissue with a negative correlation with radio sensitivity, and COL1A1 activation can inhibit the apoptosis of cervical cancer cells.27 Another study reported that COL1A1 is upregulated in muscle-invasive bladder cancer cells and knockdown of COL1A1 significantly inhibits the proliferation, migration and invasion of cancer cells by inhibiting the EMT process and the TGF-β signaling pathway.21 These studies confirm that COL1A1 plays a role as an oncogene in cancer.
To further clarify the regulatory mechanism of COL1A1 in GC development, we used GSEA pathway enrichment analysis and found that COL1A1 was significantly enriched in TGF-β signaling pathway. TGF-β can be used as a suppressor or an activator in tumorigenesis. In the early stages of tumor growth, TGF-β, as an inhibitor, can induce the expression of cyclin-dependent kinase inhibitors. Furthermore, it inhibits cell proliferation, leads to cell cycle arrest and affects cell apoptosis.18,28,29 In contrast, TGF-β acts primarily as an activator to promote tumor growth and induce invasion and metastasis by guiding epithelial and endothelial cell differentiation, promoting angiogenesis, and blocking the anti-tumor immune response in advanced tumors.30,31 Numerous experiments indicated that TGF-β regulates transcription factors and is associated with EMT.32–34 Medici et al observed that TGF-β is responsible for loss of tight junctions and a partial loss of E-cadherin, while a complete loss of E-cadherin and transformation to the mesenchymal phenotype are dependent on the Smads signaling.35 There were some researches showing the relationship between COL1A1 and the TGF-β signaling pathway in several cancers. For instance, it has been found that knockdown of COL1A1 significantly inhibits the proliferation, migration, and invasion of muscle-invasive bladder cancer (MIBC) cells by suppressing the EMT process and the TGF-β signaling pathway.21 In addition, the expression of both MRTF-A and COL1A1 can be increased through TGF-β and Wnt signaling in breast cancer.36 However, in GC, the relationship between COL1A1 and the TGF-β signaling pathway has not been elucidated. In our study, overexpression of COL1A1 promoted the expression of pSMAD3 nuclear translocation, TGFβR1 and TGFβR2 proteins, while the promoting effect was remarkably attenuated after the addition of TGF-β signaling pathway inhibitors, suggesting that overexpression of COL1A1 may promote pSMAD3 nuclear translocation thus regulating target gene transcription, resulting in pathway activation. Therefore, we found that COL1A1 promoted GC cell migration and invasion via activating the TGF-β signaling pathway. Then, we further verified the effect of the miR-133b/COL1A1/TGF-β axis on GC migration and invasion, and it was concluded that overexpression of miR-133b inhibited the EMT process in GC cells and suppressed invasion and migration of tumor cells by suppressing the TGF-β signaling pathway, which could be reversed by overexpression of COL1A1. The results illustrate the molecular mechanism by which miR-133b suppresses the invasion and migration of GC cells via the COL1A1/TGF-β axis (Figure 6). However, there are still some limitations and linearity in this study, such as lack of in vivo verification. Besides, the specific regulatory relationship between COL1A1 and TGF-β signaling pathway has not been clarified, which will be solved in our future research.
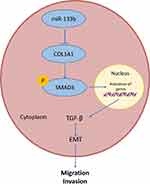 |
Figure 6 Molecular mechanism diagram of miR-133b inhibits GC cells invasion and migration through the COL1A1/TGF-β axis. |
In summary, this study demonstrates that downregulated miR-133b inhibits migration and invasion of GC cells in vitro via the COL1A1/TGF-β axis, indicating that miR-133b plays a key role in GC progression, which provides a new target for GC diagnosis and treatment.
Data Sharing Statement
The data used to support the findings of this study are included in the article. The data and materials in the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
Funding
There is no funding to report.
Disclosure
The authors declare no conflicts of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Mera RM, Bravo LE, Camargo MC, et al. Dynamics of Helicobacter pylori infection as a determinant of progression of gastric precancerous lesions: 16-year follow-up of an eradication trial. Gut. 2018;67(7):1239–1246. doi:10.1136/gutjnl-2016-311685
3. Thrumurthy SG, Chaudry MA, Chau I, Allum W. Does surgery have a role in managing incurable gastric cancer? Nat Rev Clin Oncol. 2015;12(11):676–682. doi:10.1038/nrclinonc.2015.132
4. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
5. Han X, Guo X, Zhang W, Cong Q. MicroRNA-937 inhibits the malignant phenotypes of breast cancer by directly targeting and downregulating forkhead box Q1. Onco Targets Ther. 2019;12:4813–4824. doi:10.2147/OTT.S207593
6. Bai M, Li J, Yang H, et al. miR-135b delivered by gastric tumor exosomes inhibits FOXO1 expression in endothelial cells and promotes angiogenesis. Mol Ther. 2019;27(10):1772–1783. doi:10.1016/j.ymthe.2019.06.018
7. Liu D, Zhang H, Ge S, et al. Identification of HGF as a novel target of miR-15a/16/195 in gastric cancer. Invest New Drugs. 2019. doi:10.1007/s10637-019-00834-z
8. Li X, Deng S, Pang X, et al. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int J Mol Sci. 2019;20(15):3616. doi:10.3390/ijms20153616
9. Zhao Y, Huang J, Zhang L, et al. MiR-133b is frequently decreased in gastric cancer and its overexpression reduces the metastatic potential of gastric cancer cells. BMC Cancer. 2014;14:34. doi:10.1186/1471-2407-14-34
10. Wang X, Bu J, Liu X, et al. miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget. 2017;8(38):63935–63948. doi:10.18632/oncotarget.19212
11. Zeng W, Zhu J-F, Liu J-Y, et al. miR-133b inhibits cell proliferation, migration and invasion of esophageal squamous cell carcinoma by targeting EGFR. Biomed Pharmacother. 2019;111:476–484. doi:10.1016/j.biopha.2018.12.057
12. ZiaSarabi P, Sorayayi S, Hesari A, Ghasemi F. Circulating microRNA-133, microRNA-17 and microRNA-25 in serum and its potential diagnostic value in gastric cancer. J Cell Biochem. 2019;120(8):12376–12381. doi:10.1002/jcb.28503
13. Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi:10.1038/ng1060
14. Hayashi M, Nomoto S, Hishida M, et al. Identification of the collagen type 1 α 1 gene (COL1A1) as a candidate survival-related factor associated with hepatocellular carcinoma. BMC Cancer. 2014;14:108. doi:10.1186/1471-2407-14-108
15. Oleksiewicz U, Liloglou T, Tasopoulou K-M, et al. COL1A1, PRPF40A, and UCP2 correlate with hypoxia markers in non-small cell lung cancer. J Cancer Res Clin Oncol. 2017;143(7):1133–1141. doi:10.1007/s00432-017-2381-y
16. Lv J, Guo L, Wang J-H, et al. Biomarker identification and trans-regulatory network analyses in esophageal adenocarcinoma and barrett’s esophagus. World J Gastroenterol. 2019;25(2):233–244. doi:10.3748/wjg.v25.i2.233
17. Ma H-P, Chang H-L, Bamodu OA, et al. Collagen 1A1 (COL1A1) is a reliable biomarker and putative therapeutic target for hepatocellular carcinogenesis and metastasis. Cancers. 2019;11(6):786. doi:10.3390/cancers11060786
18. Chen Y, Di C, Zhang X, et al. Transforming growth factor β signaling pathway: a promising therapeutic target for cancer. J Cell Physiol. 2020;235(3):1903–1914. doi:10.1002/jcp.29108
19. Zhang S, Gong Y, Xiao J, et al. A COL1A1 promoter-controlled expression of TGF-β soluble receptor inhibits hepatic fibrosis without triggering autoimmune responses. Dig Dis Sci. 2018;63(10):2662–2672. doi:10.1007/s10620-018-5168-3
20. Hillege MMG, Galli Caro RA, Offringa C, de Wit GMJ, Jaspers RT, Hoogaars WMH. TGF-β regulates collagen type I expression in myoblasts and myotubes via transient Ctgf and Fgf-2 expression. Cells. 2020;9(2):375. doi:10.3390/cells9020375
21. Zhu H, Chen H, Wang J, Zhou L, Liu S. Collagen stiffness promoted non-muscle-invasive bladder cancer progression to muscle-invasive bladder cancer. Onco Targets Ther. 2019;12:3441–3457. doi:10.2147/OTT.S194568
22. Zhu J-F, Liu Y, Huang H, et al. MicroRNA-133b/EGFR axis regulates esophageal squamous cell carcinoma metastases by suppressing anoikis resistance and anchorage-independent growth. Cancer Cell Int. 2018;18:193. doi:10.1186/s12935-018-0684-y
23. Huang S, Wa Q, Pan J, et al. Transcriptional downregulation of miR-133b by REST promotes prostate cancer metastasis to bone via activating TGF-β signaling. Cell Death Dis. 2018;9(7):779. doi:10.1038/s41419-018-0807-3
24. Wu Q, Zhang B, Sun Y, et al. Identification of novel biomarkers and candidate small molecule drugs in non-small-cell lung cancer by integrated microarray analysis. Onco Targets Ther. 2019;12:3545–3563. doi:10.2147/OTT.S198621
25. Tang X, Huang X, Wang D, et al. Identifying gene modules of thyroid cancer associated with pathological stage by weighted gene co-expression network analysis. Gene. 2019;704:142–148. doi:10.1016/j.gene.2019.04.017
26. Zhang X, Gao C, Liu L, et al. DNA methylation-based diagnostic and prognostic biomarkers of nonsmoking lung adenocarcinoma patients. J Cell Biochem. 2019;120(8):13520–13530. doi:10.1002/jcb.28627
27. Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17:73. doi:10.1186/s12935-017-0443-5
28. Imamura T, Hikita A, Inoue Y. The roles of TGF-β signaling in carcinogenesis and breast cancer metastasis. Breast Cancer. 2012;19(2):118–124. doi:10.1007/s12282-011-0321-2
29. Huang JJ, Blobe GC. Dichotomous roles of TGF-β in human cancer. Biochem Soc Trans. 2016;44(5):1441–1454. doi:10.1042/BST20160065
30. Muppala S, Xiao R, Krukovets I, et al. Thrombospondin-4 mediates TGF-β-induced angiogenesis. Oncogene. 2017;36(36):5189–5198. doi:10.1038/onc.2017.140
31. Zonneville J, Safina A, Truskinovsky AM, Arteaga CL, Bakin AV. TGF-β signaling promotes tumor vasculature by enhancing the pericyte-endothelium association. BMC Cancer. 2018;18(1):670. doi:10.1186/s12885-018-4587-z
32. Mani SA, Yang J, Brooks M, et al. Mesenchyme forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi:10.1073/pnas.0703900104
33. Naber HPH, Drabsch Y, Snaar-Jagalska BE, Ten Dijke P, van Laar T. Snail and slug, key regulators of TGF-β-induced EMT, are sufficient for the induction of single-cell invasion. Biochem Biophys Res Commun. 2013;435(1):58–63. doi:10.1016/j.bbrc.2013.04.037
34. Peinado H, Quintanilla M, Cano A. Transforming growth factor beta-1 induces snail transcription factor in epithelial cell lines: mechanisms for epithelial mesenchymal transitions. J Biol Chem. 2003;278(23):21113–21123. doi:10.1074/jbc.M211304200
35. Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17(4):1871–1879. doi:10.1091/mbc.e05-08-0767
36. Meng C, He Y, Wei Z, et al. MRTF-A mediates the activation of COL1A1 expression stimulated by multiple signaling pathways in human breast cancer cells. Biomed Pharmacother. 2018;104:718–728. doi:10.1016/j.biopha.2018.05.092
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.