Back to Journals » OncoTargets and Therapy » Volume 8
miR-132 can inhibit glioma cells invasion and migration by target MMP16 in vitro
Authors Wang H, Li X, Wu C, Wu Z, Li Y, Yang T, Chen G, Xie X, Huang Y, Du Z, Zhou YX
Received 15 December 2014
Accepted for publication 16 March 2015
Published 3 November 2015 Volume 2015:8 Pages 3211—3218
DOI https://doi.org/10.2147/OTT.S79282
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Daniele Santini
This paper has been retracted.
Hangzhou Wang,1,2,* Xue-tao Li,1,* Chun Wu,1 Zhi-wu Wu,1 Yan-yan Li,1 Tian-quan Yang,1 Gui-lin Chen,1 Xue-shun Xie,1 Yu-lun Huang,1 Zi-wei Du,1 You-xin Zhou1
1Department of Neurosurgery & Brain and Nerve Research Laboratory, The First Affiliated Hospital of Soochow University, Suzhou, Jiangsu, People’s Republic of China; 2Department of Neurosurgery, Children’s Hospital Affiliated to Soochow University, Suzhou, Jiangsu, People’s Republic of China
*These authors contributed equally to this work
Abstract: Gliomas are the most common malignant primary brain tumors, and new clinical biomarkers and therapeutic targets are imminently required. MicroRNAs (miRNAs) are a novel class of small non-coding RNAs (~22nt) involved in the regulation of various biological processes. Here, by using real-time polymerase chain reaction, miRNA-132 was found to be significantly deregulated in glioma tissues. Based on the prediction of the target genes of miR-132, we hypothesized that there is a significant association between miR-132 and matrix metalloproteinase (MMP) 16 (MT3-MMP), a protein of the MMP family. We showed that the up-expression of miR-132 inhibited cell migration and invasion in the human glioma cell lines A172, SHG44, and U87. Furthermore, the overexpression of miR-132 reduced the expression of MMP16 in A172, SHG44, and U87 cells. Taken together, our study suggested that miR-132 affects glioma cell migration and invasion by MMP16 and implicates miR-132 as a metastasis-inhibiting miRNA in gliomas.
Keywords: gliocytoma, microRNA, MT3-MMP, invasions
Introduction
Gliomas are the most common malignant primary brain tumors and are characterized by increased proliferation, robust angiogenesis, and invasion into the surrounding normal brain tissue.1 Despite aggressive surgery, radiation, and chemotherapy, the median survival of glioblastoma patients is 12–15 months.2 Since tumor invasion is a major reason for treatment failure,3,4 the development of novel therapeutic strategies aimed at limiting or reducing the ability of invasion of glioma cells could have a deep effect on patient outcome.
MicroRNAs (miRNAs) are short single-stranded nucleotide RNA molecules, which regulate gene expression by binding to the 3′-UTRs translated regions of their target mRNA molecules, to repress transcription or induce mRNA degradation.5,6 miRNA controls cell growth, proliferation, metabolism, and apoptosis.7,8 Indeed, specific miRNA dysregulation has been shown to correlate with particular types of cancer.9,10 For example, miR-16, lower expressed in glioma cells, suppresses Bcl-211 and miR-145, overexpressed in metastatic glioma cells, suppresses ADAM17.12
Matrix metalloproteinase (MMP) 16 (membrane type 3 MMP [MT3-MMP]) is a membrane-type metalloprotease that functions in activating proMMP2 (gelatinase A) into its active form as the zymogen is excreted out of the cell.13 Therefore, a zymogram depicting the gelatinase activity of activated MMP2 would be an indirect mechanism of determining the activity of MMP16. MMP2 can cleave collagen IV of the basement membrane and is implicated in cancer metastasis.14 It is therefore not surprising that high MMP16 expression has been associated with increasing invasiveness in gastric cancer,15 hepatocellular carcinoma,16 prostate cancer,17 as well as melanoma cells.18
hsa-miR-132 gene is located in chromosome 17 (1953202–1953302) with 100 bp in miRbase. In the last several years, many studies have found the deregulated expression of miR-132, which has been implicated in the development and progression of various cancers. For instance, miR-132 can act as a tumor suppressor and inhibit cell proliferation in breast cancer.19 Furthermore, the dysregulation of miR-132 has been found in primary osteosarcoma,20 Alzheimer’s disease,21 prostate cancer,22 and pancreatic cancer.23 But the function of miR-132 in regulating glioma cell migration and invasion remains unexplored.
In our study, the expression of miR-132 in glioma tissues compared with normal brain tissue was studied by real-time reverse transcription polymerase chain reaction (RT-PCR). We demonstrated that miR-132 inhibits glioma cell invasion by directly targeting the three prime untranslated region (3′-UTRs) of MMP16, at the time impair activation of MMP2. Our results suggest that downregulation of miR-132 plays an important role in enhancing the invasion of glioma cells.
Materials and methods
Cell lines and cell culture
The following three human glioma cell lines were used: U87MG, SHG44, and A172. They were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (U87MG, A172) or RPMI-1640 (SHG44) medium supplemented with 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin. All cultures were maintained in a humidified atmosphere of 5% CO2 at 37°C.
Clinical specimens
Glioma tissues were obtained from therapeutic procedures performed as routine clinical management at our institution. Tissue samples were resected during surgery and immediately frozen in liquid nitrogen for subsequent total RNA extraction. A total of eleven low-grade glioma tissues (glioma grades I–II), 12 high-grade glioma tissues (glioma grades III–IV), and eight nonneoplastic brain specimens were included in our study.
Transfection of miRNA mimics or inhibitor
Glioma cells were seeded in six-well plates at 50% confluence without antibiotics on the day before transfection. Transfection with miR-132 mimics (UAACAGUCUACAGCCAUGGUCGACCAUGGCUGUAGACUGUUAUU) or miRNA mimics negative control (UUCUCCGAACGUGUCACGUTTACGUGACA CGUUCGGAGAATT) and miR-132 inhibitor (5′-3′CGACCAUGGCUGUAGACUGUUA) or miRNA inhibitor negative control (5′-3′CAGUACUUUUGUGUAGUACAA) was performed using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). Transfection complexes were prepared according to the manufacturer’s instructions.
RNA extraction and quantitative real-time PCR
Total RNA was isolated from tumor sample and glioma cell lines with TRIzol reagent (Thermo Fisher Scientific), and their RNA concentrations were measured with an Eppendorf BioPhotometer at 260 nm and 280 nm (A260/280). cDNA was synthesized with the ReverTra Ace®qPCR RT kit (FSQ-101; Toyobo, Osaka, Japan). Real-time PCR analyses were performed with SYBR® Green Real-Time PCR Master Mix (QPK-201; Toyobo). Quantification of miR-132 was performed with a stem-loop real-time PCR miRNA kit (RiboBio, Guangzhou, People’s Republic of China). Operation is based on manufacturer’s instructions. The primer pairs for MMP16 were sense 5′-GAAGACGGTTGGATTTCGTG-3′ and antisense 5′-GTCAGTCGGTGGAAGGTAGC-3′, and those for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were sense 5′-GGGTGTGAACCATGAGAAGT-3′ and antisense 5′-GGCATGGACTGTGGTCATGA-3′. RT-PCR was done in a preheated real-time instrument (ABI7300) for 40 cycles of 15 seconds at 94°C and of 1 minute at 58°C.
In vitro matrigel invasion assay
Matrigel chambers (BD Biosciences) were used to determine the effect of miR-132 on invasiveness according to the manufacturer’s instructions. A total of 5×104 cells, after being transfected 36 hours, were resuspended in basic DMEM culture medium, supplemented with 0.5% FBS, and added 200 μL to the upper chamber of a transwell system (8 μm pore size, Corning 3422), while the lower chamber was filled with 0.5 mL of complete medium that served as a chemoattractant. After incubation for 24 hours at 37°C, invasive cells, which had the ability to push themselves through the 8 μm pores and grow on the lower surface, were fixed with 100% methanol and stained with 1% toluidine blue (Sigma-Aldrich Co.) before counting under an inverted microscope. All the experiments were done in duplicate, and results were expressed as mean ± SEM of three independent experiments.
Western blotting
Forty-eight hours after miRNA mimics or miRNA inhibitor transfection, total proteins were isolated from tissues and cell lines with radioimmunoprecipitation (RIPA) lysis buffer (P0013B, Beyotime, People’s Republic of China). The supernatants containing the whole protein extracts were obtained after centrifugation of the lysates at 12,000× g for 20 minutes at 4°C. The protein concentrations were determined by enhanced bicinchoninic acid protein assay kit (Beyotime). Heat-denatured protein samples (50 μg per lane) were resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE) gel and transferred to an Immobilon-P transfer membrane (EMD Millipore). The membrane was blocked in 5% non-fat dry milk in Tris-buffered saline (pH 7.4) containing 0.05% Tween-20 to block nonspecific binding, followed by incubation overnight at 4°C with a primary rabbit polyclonal antibody against human MMP16 (1:200, Boster, People’s Republic of China), MMP2 (1:250, Santa Cruz Biotechnology Inc., Dallas, TX, USA), and was blotted with goat anti-rabbit immunoglobulin G (1:3,000, Santa Cruz Biotechnology Inc., USA). Then GAPDH was used as a loading control. Signals were detected by secondary antibodies labeled with HRP, and the bound antibody was detected with the use of enhanced chemiluminescence detection reagents (Beyotime) according to the manufacturer’s instructions.
Plasmids construction and dual-luciferase reporter assay
We entrusted Shanghai Ltd. to construct pGL3-MMP16 by amplifying 3′-UTR of MMP16 gene harboring the miR-132 binding site predicted by the TargetScan (http://www.targetscan.org) and subsequently cloning it into the pGL3 control vector (Promega Corporation, Fitchburg, WI, USA) at the Xbal site immediately downstream of firefly luciferase. pGL3-MMP16-mut, which has three mismatch mutations in the miR-132 seed complementary site, was generated to be a negative control. For the luciferase assay, 293T cells were cultured in 12-well plates and each was cotransfected with 400 ng of either pGL3-MMP16 or pGL3-MMP16-mut, 50 ng of pRL-TK (Promega Corporation), and 50 nmol/L of miR-132 mimics or NC. The pRL-TK Renilla luciferase plasmid was used as an internal control to correct differences in both transfection and harvest efficiencies.
Forty-eight hours after transfection, firefly and Renilla luciferase activities were measured using the dual-luciferase reporter assay (Promega Corporation). The results were expressed as relative luciferase activity (firefly luciferase/Renilla luciferase).
Statistical analysis
Data are presented as mean ± standard deviation. The data were analyzed using the SPSS 12.0 Windows version software. Statistical analyses were done by analysis of variance or Student’s t-test. P-value <0.05 was considered statistically significant.
Result
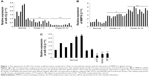
Expression of miR-132 is lower in glioma tissue compared to normal brain tissues
To determine the role of miR-132 in glioma tissues, we used real-time PCR analyses to determine the expression of miR-132 in eleven low-grade glioma tissues and 12 high-grade glioma tissues compared to eight normal brain tissues. As shown in Figure 1A, miR-132 expression levels were significantly decreased in glioma tissues compared to normal brain tissues.
In addition, we found that the expression of miR-132 was lower in glioma cell lines compared to normal brain tissues (Figure 1B). The downregulation of miR-132 in glioma tissues and glioma cell lines suggests that miR-132 may be as a potential target in glioma therapy.
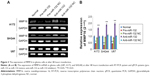
miR-132 can inhibit invasion of glioma cells
In order to understand the effect of miR-132 on glioma cell invasion and migration, we used miR-132 mimics and inhibitors to infect human glioma U87, SHG44, and A172 cells. Seventy-two hours after transfection with miR-132 or scrambled-miRNA, U87, SHG44, and A172 cells were seeded into the upper chamber, and then cells that invaded through the extracellular matrix (ECM) after 48 hours were imaged and counted (Figure 2A and B). In both cell lines, miR-132 could decrease the number of cells that invaded compared to controls. Taken together, these data indicate that miR-132 may be as a regulatory molecule concern cell migration and invasion in vitro.
Expression of MMP16 mRNA in glioma tissues
Real-time PCR analyses showed that the expression of MMP16 mRNA was dramatically higher in eleven low-grade glioma tissues and 12 high-grade glioma tissues compared to eight normal brain tissues. The result is shown in Figure 1C. Here, our result showed that MMP16 mRNA is higher expression in glioma tissues, compared that the results of miR-132 in glioma tissues suggest that the expression of MMP16 mRNA expression is inversely related to the miR-132.
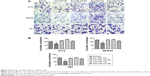
miR-132 downregulates the expression of MMP16 mRNA
To explicit the mechanism by which miR-132 inhibited cell migration in glioma cells, we founded miR-132 targets using the algorithms TargetScan5 and miRBase and found that MMP16 might play a role in cell migration and invasion. We use RT-PCR assay and qPCR systems. Transfection of glioma cells with miR-132 mimic significantly decreased the expression of MMP16 mRNA in glioma cells compared to levels in control miR expressing cells (Figure 3A and B). The above results showed that MMP16 may be a target gene of miR-132 to promote the glioma cell migration and invasion.
MMP16 as an activator of MMP2 can activate the expression of MMP2 protein levels
To validate whether miR-132 can affect the expression levels of MMP16 protein, we measured the protein expression levels of MMP16 in response to the effect in the miR-132 expression level in U87, A172, and SHG44 by transfection. We found that protein expression level of MMP16 was downregulated and upregulated by miR-132 mimics and miR-132 inhibitor transfection, respectively (Figure 4).
miR-132 can directly downstream the target gene of MMP16
miR-132 is predicted to target MMP16 (MT3-MMP) (www.microRNA.org), which is a potential activator of MMP2.23,24 We used Western blot to validate the hypothesis. The results showed that miR-132 can activate the expression of MMP2 protein by regulating the expression of MMP16 (Figure 4). Up-miR-132 inhibits invasion in human glioma cells by directly downregulating MMP16 expression. The luciferase assay revealed reduced relative luciferase activities in 293T cells stably, overexpressing miR-132 following transfection of MMP16 3′-UTR (Figure 5A and B) (P<0.05).
Discussion
In our study, we found lower levels of miR-132 in glioma cancer tissues than in normal brain tissues and in glioma cells than in normal brain tissues. Furthermore, we have found that miR-132 could inhibit glioma cell invasion and migration in glioma cells. Glioma cells by transfected miR-132 mimics decreased tumor cell migration and invasion, while inhibition of transfecting miR-132 inhibitor produced the opposite result. Nevertheless, whether miR-132 inhibits tumor invasion and metastasis of glioma in vivo still remains to be investigated, because such study is hampered at present by the lack of an experimental strategy for stably increasing or silencing miRNAs over extended periods of time.
We further showed that the function of miR-132 inhibits glioma cell invasion and migration properties by reducing the expression of an MMP gene, MMP16. MMP16 is a membrane-anchored MMP that is able to activate MMP2 for invasion. Our study suggests the functional relevance of MMP16 in the downregulation of glioma cell migration. The previous report showed that decreasing MMP16 levels efficiently inhibit cell invasion of glioma cells.25 As MMP2 is activated by MMP16, this would be an indirect indication of the function of MMP16. MMP16 was also found to possess proteolytic activity against ECM components such as type III collagen. It is possible that MT3-MMP is involved in the turnover of ECM in the normal brain and astrocytic tumor tissues.26,27
In conclusion, our data suggested that miR-132 plays a key role in the malignancy of glioma cells possibly by direct regulation of MMP16 protein expression, which affects glioma cell migration and invasion. Our data suggested that miR-132 may be a potential therapeutic target for preventing GBM invasion and metastasis. However, further study is needed to determine if MMP16 activity is influenced by miR-132 in vivo in glioma.
Acknowledgments
We thank our central lab for providing technical instruction and assistance. This work was partly supported by National Science Foundation (Grant No 81372689) and the Foundation of Health Department in Jiangsu Province (Grant No K201106).
Disclosure
The authors report no conflicts of interest in this work.
References
Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. | ||
Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. | ||
Giese A, Bjerkvig R, Berens ME, Westphal M. Cost of migration: invasion of malignant gliomas and implications for treatment. J Clin Oncol. 2003;21:1624–1636. | ||
Lefranc F, Brotchi J, Kiss R. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J Clin Oncol. 2005;23:2411–2422. | ||
Stupp R, Mason WP, van den Bent MJ, et al; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. | ||
Holland EC. Gliomagenesis: genetic alterations and mouse models. Nat Rev Genet. 2001;2:120–129. | ||
Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–568. | ||
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. | ||
Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. | ||
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. | ||
Yang TQ, Lu XJ, Wu TF, et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci. 2014;105(3):265–271. | ||
Yong Lu, Michael Chopp, Xuguang Zheng, Mark Katakowski, Benjamin Buller, Feng Jiang. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncology Reports. 2013; 29:67–72. | ||
Nakada M, Nakamura H, Ikeda E, et al. Expression and tissue localization of membrane-type 1, 2, and 3 matrix metalloproteinases in human astrocytic tumors. Am J Pathol. 1999;154:417–428. | ||
Rowe RG, Weiss SJ. Breaching the basement membrane: who, when and how? Trends Cell Biol. 2008;18:560–574. | ||
Lowy AM, Clements WM, Bishop J, et al. Beta-Catenin/Wnt signaling regulates expression of the membrane type 3 matrix metalloproteinase in gastric cancer. Cancer Res. 2006;66:4734–4741. | ||
Arai I, Nagano H, Kondo M, et al. Overexpression of MT3-MMP in hepatocellular carcinoma correlates with capsular invasion. Hepatogastroenterology. 2007;54:167–171. | ||
Daja MM, Niu X, Zhao Z, Brown JM, Russell PJ. Characterization of expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in prostate cancer cell lines. Prostate Cancer Prostatic Dis. 2003;6:15–26. | ||
Ohnishi Y, Tajima S, Ishibashi A. Coordinate expression of membrane type-matrix metalloproteinases-2 and 3 (MT2-MMP and MT3-MMP) and matrix metalloproteinase-2 (MMP-2) in primary and metastatic melanoma cells. Eur J Dermatol. 2001;11:420–423. | ||
Shuai Li, Huimin Meng, Feng Zhou, et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathology – Research and Practice. 2013;179–183. | ||
Yang J, Gao T, Tang J, Cai H, Lin L, Fu S. Loss of microRNA-132 predicts poor prognosis in patients with primary osteosarcoma. Mol Cell Biochem. 2013;381(1–2):9–15. | ||
Xia H, Qi Y, Ng SS, et al. MicroRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 2009;1269:158–165. | ||
Takino T, Sato H, Shinagawa A, Seiki M. Identification of the second membrane-type matrix metalloproteinase (MT-MMP-2) gene from a human placenta cDNA library. MTMMPs form a unique membrane-type subclass in the MMP family. J Biol Chem. 1995;270:23013–23020. | ||
Nakada M, Yamada A, Takino T. Suppression of membrane-type 1 matrix metalloproteinase (MMP)-mediated MMP-2 activation and tumour invasion by testican 3 and its splicing variant gene product, N-Tes. Cancer Res. 2001;61:8896–8902. | ||
Li S, Meng H, Zhou F, et al. MicroRNA-132 is frequently down-regulated in ductal carcinoma in situ (DCIS) of breast and acts as a tumor suppressor by inhibiting cell proliferation. Pathol Res Pract. 2013;209:179–183. | ||
Li Y, Wang Y, Yu L, et al. miR-146b-5p inhibits glioma migration and invasion by targeting MMP16. Cancer Lett. 2013;339(2):260–269. | ||
Fillmore HL, VanMeter TE, Broaddus WC. Membrane-type matrix metalloproteinases (MT-MMPs): expression and function during glioma invasion. J Neurooncol. 2001;53(2):187–202. | ||
Lu Y, Chopp M, Zheng X, Katakowski M, Buller B, Jiang F. MiR-145 reduces ADAM17 expression and inhibits in vitro migration and invasion of glioma cells. Oncol Rep. 2013;29:67–72. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.