Back to Journals » Cancer Management and Research » Volume 13
miR-130a-Mediated KLF3 Can Inhibit the Growth of Lung Cancer Cells
Authors Wei MC, Wang YM, Wang DW
Received 26 October 2020
Accepted for publication 8 March 2021
Published 6 April 2021 Volume 2021:13 Pages 2995—3004
DOI https://doi.org/10.2147/CMAR.S281203
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Eileen O'Reilly
Ming-Chao Wei,1 Yu-Min Wang,2 Da-Wei Wang3
1Department of Thoracic Surgery, Yantai Affiliated Hospital of Binzhou Medical University, Shandong, People’s Republic of China; 2Binzhou Medical University, Shandong, People’s Republic of China; 3Department of Thoracic Surgery, Yantai Mountain Hospital, Shandong, People’s Republic of China
Correspondence: Da-Wei Wang
Department of Thoracic Surgery, Yantai Mountain Hospital, Yantai, Shandong, 264001, People’s Republic of China
Tel +86-0535-6602183
Email [email protected]
Background: The role of microRNA (miR) in tumors has been reported in numerous articles. Previous studies have found that miR-130a is low expressed in lung cancer, but the related mechanism has not been fully elucidated. This study mainly explores the mechanism of miR-130a in lung cancer, so as to provide potential therapeutic targets for clinical applications.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression of miR-130a and KLF3 in the tissues of lung cancer patients. The miR-130a-mimics and miR-130a-inhibit were constructed. Cell proliferation, invasion, migration and apoptosis were determined by CCK-8, transwell, scratch test and flow cytometry. Western Blot was used to determine the expression of KLF3 protein in cells, and the dual-luciferase reporter to determine the relationship between KLF3 and miR-130a.
Results: miR-130a shows low expression in NSCLC patients, while KLF3 shows high expression, exhibiting a negative correlation. The 5-year survival rate of patients with low miR-130a expression and high KLF3 expression was reduced. Cox regression analysis showed that miR-130a was an independent prognostic factor for NSCLC patients. The dual-luciferase reporter revealed that miR-130a bound to KLF3 in a targeted manner, and cell experiments showed that miR-130a could inhibit the growth of lung cancer cells by regulating the expression of KLF3.
Conclusion: miR-130a shows low expression in lung cancer and predicts a poor prognosis. In addition, up-regulation of miR-130a can down-regulate KLF3 and inhibit the growth of lung cancer.
Keywords: miR-130a, KLF3, lung cancer, prognosis
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with the highest mortality and morbidity.1 Statistics show that more than 2 million new cases of lung cancer patients have been diagnosed in 2018, with 1.7 million deaths. The incidence of lung cancer is on the rise year by year.2,3 Lung cancer can be divided into small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) according to pathological tissues, with NSCLC accounting for more than 80% of lung cancer.4,5 Surgical resection is the optimal treatment for early-stage NSCLC. However, most of the patients can only improve their condition through radiotherapy and chemotherapy as the disease has already progressed to the middle and advanced stage after diagnosis. Although chemoradiotherapy can improve the patient’s condition, its prognosis is poor, and the disease is prone to recurrence.6,7 Therefore, it is urgent to better understand the pathogenesis of lung cancer. With the development of scientific research in recent years, it has been found that the occurrence of NSCLC is associated with gene silencing of tumor suppressor genes.
microRNA (miR) is a newly discovered type of eukaryotic cell widely found in recent years. It is a highly conserved endogenous non-coding short-chain nucleotide transcript with a size between 18 and 25 nucleotides. It can be combined with the target gene 3ʹ UTR through complementary pairing, so as to degrade or inhibit the expression of the target gene.8–10 As a member of the miRs family, studies have found that miR-130a shows low expression in a variety of tumors, such as liver cancer, breast cancer and ovarian cancer.11–13 Moreover, other studies suggested that miR-130a could further inhibit tumors by regulating downstream target genes. For example,14 miR-130a can affect the angiogenesis of gastric cancer by regulating C-MYB. Besides, there is a study supporting that miR-130a can inhibit proliferation, migration and invasion of liver cancer cells through down-regulating Rho-kinase 2.15 However, there are few studies on the relationship between miR-130a and NSCLC. Previous studies have found low expression of miR-130a in lung cancer, while the relevant mechanism has not been fully elucidated.
Therefore, in this study, we analyzed the related mechanism of miR-130a in NSCLC to provide potential therapeutic targets for clinical use.
Materials and Methods
Data
A total of 50 cases of patients with lung cancer receiving treatment in our hospital from May 2012 to June 2014 were collected as research objects of this study. During the operation, patients’ cancer tissues and para-carcinoma tissues were collected, transported by liquid nitrogen, and stored at −80°C. Inclusion criteria: patients were diagnosed as NSCLC by pathology or iconography; patients knew about the study and signed the informed consent; patients’ symptoms were in line with the 8th edition of NSCLC staging standard issued by Union for International Cancer Control (UICC).16 Exclusion criteria: patients were complicated with other tumors; patients did not cooperate with follow-up; patients’ expected survival was longer than 1 month; patients received anti-tumor treatment before this study. This study was approved by the ethical committee of Yantai Mountain Hospital. The patients were followed up by telephone and outpatient clinic to review the electronic pathological records. This study was fully consistent with the Declaration of Helsinki.
Cell Culture and Transfection
Lung cancer cells H1299, H1975, H2228, A549 and human bronchial epithelial cells BEAS-2B were purchased from American Type Culture Collection (ATCC, Manassas, Virginia, USA). All cells were cultured in dulbecco’s modified eagle medium (DMEM) with 10% fetal bovine serum (FBS, Sigma, St. Loius) at 37°C with 5% CO2. miR-130a mimics, miR-130a-inhibit and negative controls (miR-NC/miR-inhibit) were designed and synthesized by RiboBio Co., Ltd. (Guangzhou, China). Krüppel-like factor 3 (KLF3) over-expression (pcDNA3.1-KLF3), inhibitor (si-KLF3), and negative control (si-NC) were also constructed with pcDNA3.1 as a vector. The Lipofectamine 3000 kit (Invitrogen) was used for transfection. The transfection steps were performed in strict accordance with the kit instructions.
qRT-PCR Detection
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the instructions of manufacturer. The TaqMan miRNA reverse transcription kit (Applied Biosystems) was used to quantify miRNA expression by quantitative real-time polymerase chain reaction (qRT-PCR), and cDNA obtained was utilized for further study. The PrimeScript RT Master Mix kit was utilized for PCR amplification (Takara Bio, Japan). The amplification system and amplification conditions were prepared in accordance with the kit instructions. The ABI 7500 was utilized in the experiment. Three parallel duplicate wells were designed in the experiment, and all samples were tested three times. GADPH was used as internal reference for GENE, and U6 as internal reference for miR. 2−ΔΔct was applied to analyze the data.17 Primer sequence of miR-130a: forward primer: 5ʹ-TTCACATTGTGCTACTGTCTGC-3ʹ, reverse primer: 5ʹ-GCTCTGACTTTATTGCACTACT-3ʹ. Primer sequence of KLF3: forward primer: 5ʹ-TGTCTCAGTGTCATACCCATCT-3ʹ, reverse primer: 5ʹ-CCTTCTGGGGTCTGAAAGAACTT-3ʹ. Primer sequence of U6: forward primer: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, reverse primer: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. Primer sequence of GADPH: forward primer: 5ʹ-AGAAGGCTGGGGCTCATTTG-3ʹ, reverse primer: 5ʹ-AGGGGCCATCCACAGTCTTC-3ʹ.
Western Blot Detection
The total protein was extracted from each group of cells after collection of culture using the RIPA lysis method, and the protein concentration was measured using BCA assay. The protein concentration was adjusted to 4 μg/μL, separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF) membrane. The membrane was then stained with Ponceau S, washed after soaking in PBST for 5 min, and blocked with 5% skimmed milk powder for 2 h. Then, KLF3 primary antibody (1:500, abcam, USA) was added for sealing at 4°C for one night. The primary antibody was removed by washing membrane, and horseradish peroxidase labeled goat anti-rabbit secondary antibody (1:1000, abcam, USA) was added for incubation at 37°C for an hour, and then rinsed with PBS for 3 times, with 5 min each. Excess liquid was absorbed from the membrane with filter paper. The enhanced chemiluminescence (ECL) was used for illumination and development in a dark room. The protein bands were scanned, and the grayscale value was analyzed in Quantity One software. The relative expression level of protein = grayscale value of the target protein band/grayscale value of β-actin protein band.
Cell Counting Kit-8 (CCK-8) Experiment
Cells after 48 h of transfection were collected for detection using the CCK-8 kit (med chem express, USA). The specific scheme was as follows: cells were adjusted to 1*105 and transferred to 96-well plates. The CCK-8 reagent was added to each well at 6 h, 24 h, 48 h and 72 h, respectively, to incubate at 37°C for 30 min. The absorbance was detected at 450 nm with a microplate reader.
Transwell Experiment
The transfected cells were collected and transferred to 24-well Transwell plates (8-μm wells, Corning, New York, USA). Matrigel matrix (BD Biosciences, San Diego, California, USA) was smeared on the plate. Cells were adjusted to 5 x 104/mL and suspended in 200 μL DMEM (containing 1% FBS) on the upper compartment of Transwell, and the lower compartment was added with medium containing 15% FBS. After 48 h of culture, cells were fixed with 10% methanol for 15 min and washed three times with double distilled water. After it was dried, it was stained with 0.5% crystal violet, and the cell invasion was observed with a microscope.
Scratch Test
After transfection, the cells were spread out in 6-well plates. When the cell monolayer reached 90% confluence, a 10 μL pipette tip was used to wound. Then, the cells were cultured in a medium containing 1% FBS for 24 h, and the healing conditions were observed under an inverted phase contrast microscope.
Annexin V-FITC/PI Analysis
Transfected cells were collected and digested with 0.25% trypsin (Gibco). After digestion, they were washed twice with PBS and added with 100 μL binding buffer. They were configured into 1*106/mL suspension, and Annexin V-FITC and PI (BestBio, Shanghai, China) were successively added. The cells were incubated at room temperature in the dark for 5 min. The FC500MCL flow cytometry system (Beckman Coulter, USA) was used for determination, and the experiment was repeated three times to take the average value.
Dual-Luciferase Reporter
Dual-luciferase report was adopted to determine the relationship between miR-130a and KLF3. With psi-CHECK2 as the vector, KLF3 wild type (KLF3-WT) and mutant (KLF3-MUT) sequence were constructed, respectively. They were co-transfected with miR-130a-mimics and miR-130a-inhibit into 293T cells, respectively. After 48 h after transfection, the activity of firefly and renin luciferase in the cell lysates was continuously determined using the dual-luciferase report kit (Promega, USA).
Tumor Formation Experiment in Nude Mice
BALB/C nude mice (10 males, 4–6 weeks old) were purchased from Vital River, Beijing, China. All animal experiments were conducted in accordance with the Guidelines for Welfare and Ethical Review of Laboratory Animal. The lung cancer xenograft model was established by suspending 5×106 miR-130a-mimics-transfected A549 cells in 200 μL phosphate buffer and then subcutaneously inoculating the solution into the right abdomen of nude mice. The length (L) and width (W) were measured with vernier caliper every 7 days to monitor tumor size, and the volume was calculated using the formula (L×W 2)/2. On the 28th day, the nude mice were euthanized to collect tumor tissues. The study was approved by the medical Ethics Committee of Yantai Mountain Hospital, and was in accordance with the Laboratory animal—Guideline for ethical review of animal welfare.
Statistical Analysis
In this study, GraphPad 7 software package was used to draw the required images and analyze the data, and SPSS20.0 software package to analyze the independent prognostic factors of patients. The data was represented by mean ± standard deviation (Mean±SD). Intergroup comparison was performed by independent sample t-test. Enumeration data was represented using percentage (%), and chi-square test was adopted and represented by χ2. Comparison between groups was analyzed using one-way analysis of variance, represented with F. LSD t-test was used for afterwards pairwise comparison. Comparison among multiple time point expression was analyzed using Repeated Measures, denoted by F. Bonferroni was used for back testing. Correlations between genes were analyzed using Pearson test. The overall survival of patients was plotted using KM survival curve, and Log-rank test was used for analysis. Multivariate Cox regression analysis was applied to analyze the prognosis of patients. When P<0.05, there was statistical differences.
Results
miR-130a is Poorly Expressed in NSCLC Patients with Poor Prognosis
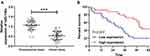
In this study, we first determined the expression of miR-130a in NSCLC patients by qRT-PCR, and the results showed that miR-130a expression was significantly lower in cancer tissues than in paracancerous tissues (Figure 1A). We then divided the patients into a high expression group and a low expression group according to the relative median expression value of miR-130a and analyzed the differences in clinical data between the two groups. Through analysis, it was found that patients with low expression were significantly more prone to lymphatic metastasis and high TNM stage (Table 1). In addition, survival analysis revealed a significantly lower 5-year survival rate in patients with low miR-130a expression (Figure 1B), and Cox regression analysis found that miR-130a was an independent factor affecting patient prognosis (Table 2).
 |
Table 1 Relationship Between miR-130a and the Pathological Data of NSCLC |
 |
Table 2 Prognostic Analysis of NSCLC Patients |
Up-Regulation of miR-130a Can Inhibit the Growth and Migration of Lung Cancer Cells
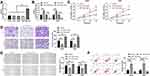
Through determination, it was found that miR-130a expression in lung cancer cells was significantly lower than that in human lung bronchial epithelial cells (Figure 2A). To observe its effects on lung cancer cells, we transfected differentially expressed miR-130a into cells (Figure 2B). CCK-8 test showed compared to miR-NC, cell proliferation ability was inhibited more obviously after transfection of miR-130a-mimics (Figure 2C). Transwell, Scratch test and flow cytometry experiments revealed that compared to miR-NC, transfection of miR-130a-mimics significantly inhibited cell membrane penetration and healing, and induced cell apoptosis (Figure 2D–F). In addition, cell biological function was reversed after miR-130a knock down.
miR-130a Can Target KLF3
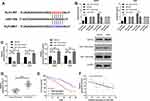
To further determine the reverse target gene of miR-130a, we found KLF3 through prediction websites of starBase, TargetScan, and miRDB (Figure 3A). To verify the relationship between miR-130a and KLF3, the dual-luciferase report was applied to detect the expression of KLF3 in lung cancer cells after transfection of miR-130a-mimics/inhibit (Figure 3B). The results showed that miR-130a-mimics inhibited the luciferase activity of KLF3-WT, while miR-130a-inhibit increased the luciferase activity of KLF3-WT (Figure 3B). Further assays showed that the expression of KLF3 mRNA and protein was significantly decreased in lung cancer cells after transfection of miR-130a-mimics compared to miR-NC, and the expression of KLF3 mRNA and protein was significantly increased in lung cancer cells of miR-130a-inhibit (Figure 3C). In addition, we also detected KLF3 expression in patient tissues and found that KLF3 expression was significantly higher in cancer tissues than in paracancerous tissues, and the 5-year survival rate was reduced in patients with high expression (Figure 3D–E). Correlation analysis revealed a negative correlation between KLF3 and miR-130a (Figure 3F).
miR-130a Targets KLF3 to Inhibit Lung Cancer Cell Growth
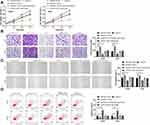
In these studies, we determined the relationship between miR-130a and KLF3. To observe the role of miR-130a and KLF3 in lung cancer, we constructed co-transfected cells and established pcDNA3.1-KLF3, si-KLF3, pcDNA3.1-KLF3+miR-130a-mimics, and si-KLF3+miR-130a separately to inhibit miR-NC+si-NC, so as to further observe the cell biological functions. The results showed that the transfection of pcDNA3.1-KLF3 accelerated the proliferation, invasion and migration of lung cancer cells, and reduced apoptosis, whereas the opposite result was observed after transfection of si-KLF3 (Figure 4A–C). In addition, after co-transfection, miR-130a-mimics could inhibit the proliferation, invasion and migration of pcDNA3.1-KLF3 on lung cancer cells and reduce apoptosis (Figure 4D), and the co-transfection outcomes of si-KLF3 and miR-130a-inhibitors were consistent with this above.
Up-Regulation of miR-130a Inhibits Tumor Growth in Nude Mice
At the end of the study, we established nude mice model of lung cancer to analyze the role of miR-130a/KLF3 axis in lung cancer. The results showed that the tumor volume of nude mice transfected with miR-130a-mimics decreased gradually with time compared with those transfected with miR-NC. After 28 days, the nude mice were executed to collect tumor tissues. It was found that the tumor tissues were significantly reduced in tumor mass after transfection of miR-130a-mimics when compared with miR-NC (Figure 5A). qRT-PCR and WB experiments revealed that the expression of miR-130a was increased and the expression of KLF3 mRNA and protein expression was significantly decreased in the tumor tissues of nude mice in miR-130a-mimics group (Figure 5B–C), suggesting that the miR-130a/KLF3 axis is involved in the development of lung cancer and is expected to be a potential therapeutic target.
Discussion
Lung cancer is a malignant tumor with the highest incidence in clinical practice and has the characteristics of easy recurrence and metastasis.18 Since most patients have advanced to the middle and late stage after diagnosis, they have missed the optimal treatment period and can only improve their conditions through chemoradiotherapy.19 Although the patient’s condition was improved after chemoradiotherapy, their prognosis is still poor.20 Therefore, it is urgent to find out the relevant mechanism of NSCLC.
miR is a short non-coding RNA that has previously been found to express in a variety of tumors and participate in tumor progression. For example, over-expression of microRNA-30a-5p was found to inhibit the proliferation and induce apoptosis of liver cancer cells by targeting the MTDH/PTEN/AKT pathway.21 miR was also found to be a potential prognostic indicator for colon cancer as well as a drug sensitization target.22 miR-130a is located on human chromosome 11q12.1. There have been few previous studies on lung cancer. Although some studies have confirmed the low expression of miR-130a in lung cancer, the relevant mechanism remains unclear.23 In this study, we found that miR-130a was low expressed in lung cancer patients and decreased in lung cancer cells, which is consistent with the study of the team of Lin.24 In addition, our further studies revealed that patients with low miR-130a expression had significantly higher TNM staging and incidence of lymphatic metastasis, suggesting that miR-130a could be used as a potential diagnostic indicator for NSCLC staging and lymphatic metastasis. However, normal samples were not collected in this study and there is room for improvement. Our further analysis revealed that the 5-year survival rate of patients with low miR-130a expression was significantly reduced, and Cox regression analysis also indicated that patients with high miR-130a expression had an increased probability of poor prognosis, which was a prognostic indicator of NSCLC. Through the above studies, it can be confirmed that miR-130a is involved in the development of lung cancer, but its function in lung cancer remains unclear.
To determine the role of miR-130a in lung cancer, we constructed miR-130a-mimics and miR-130a-inhibit to assess the cell biological function and observe the effects of miR-130a on lung cancer through transfection. It could be seen from the results that up-regulation of miR-130a led to significantly inhibited cell proliferation, invasion and migration, as well as induced apoptosis. However, after down-regulation of miR-130a, the results were reversed, the proliferation, invasion and migration of cells were significantly accelerated, and apoptosis was inhibited. Such above suggests that direct regulation of miR-130a can affect the course of lung cancer cells, and that miR-130a is a potential therapeutic target for the treatment of lung cancer.
In recent years, more and more studies have shown that miRs inhibit lung cancer growth through regulating downstream target genes. For example, miR-195 targets CHEK1 to inhibit NSCLC.25 Another study supports that miR-340 inhibits the proliferation of NSCLC tumor cells and induces the apoptosis by targeting multiple p27 negative regulatory factors.26 To further confirm the mechanism of miR-130a, we predicted its downstream target genes and did found the targeted binding sites between KLF3 and miR-130a through online sites. KLF3 is one of the 18 members of the KLF DNA-binding protein family, which is responsible for controlling gene expression at transcription levels in a variety of biological environment.27 It is reported that KLF3 is an important regulator of lung cancer metastasis by regulating the expression of STAT3 in lung cancer.28 In order to verify the expression of KLF3 in lung cancer, we performed determination. It was found that KLF3 was highly expressed in cancer tissues of NSCLC patients, and the 5-year survival rate of patients with high expression was reduced. What is more, correlation analysis revealed a negative relationship between KLF3 and miR-130a. Therefore, we speculated that miR-130a could affect the occurrence of lung cancer through KLF3.
To verify the conclusion of our research, we applied dual-luciferase reporter and found that KLF3 was regulated by miR130a. Furthermore, we determined lung cancer cells transfected with miR-130a-mimics/inhibit and found that KLF3 expression in lung cancer cells was inhibited after miR-130a up-regulation, whereas the transfection of miR-130a-inhibit had the opposite effects. At the end of the study, we conducted rescue experiments to determine the effects of miR-130a on the growth of lung cancer cells by regulating KLF3. The results showed that the proliferation, invasion and migration of cells were accelerated and the inhibition of apoptosis rate was reversed through co-transfection of miR-130a-mimics, which further confirms our findings.
There are some shortcomings in this study. First of all, we did not collect other samples, such as bronchoalveolar lavage fluid, serum, peripheral blood mononuclear cells and exosome to verify the diagnostic value of miR-130a in lung cancer. Next, we did not conduct in vivo experiments (tumor allograft implantation) to observe the effects of miR-130a on tumor in vivo. Therefore, we hope to collect more clinical samples in future studies and carry out more basic studies to supplement our findings.
In conclusion, miR-130a shows low expression in lung cancer and predicts a poor prognosis. Moreover, up-regulation of miR-130a can down-regulate KLF3 and inhibit lung cancer growth.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi:10.1007/978-3-319-24223-1_1
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Chen WQ, Zuo TT, Zheng RS, Zeng HM, Zhang SW, He J. [Lung cancer incidence and mortality in China in 2013]. Chin J Oncol. 2017;39(10):795–800. doi:10.3760/cma.j.issn.0253-3766.2017.10.015.Chinese.
4. Yu T, Zhong D. [Clinical development of immunotherapy for small cell lung cancer]. Chin J Lung Cancer. 2018;21(12):918–923. doi:10.3779/j.issn.1009-3419.2018.12.10.Chinese.
5. Nanavaty P, Alvarez MS, Alberts WM. Lung cancer screening: advantages, controversies, and applications. Cancer Control. 2014;21(1):9–14. doi:10.1177/107327481402100102
6. Yan X, Jiao SC, Zhang GQ, Guan Y, Wang JL. Tumor-associated immune factors are associated with recurrence and metastasis in non-small cell lung cancer. Cancer Gene Ther. 2017;24(2):57–63. doi:10.1038/cgt.2016.40
7. Usuda K, Sagawa M, Motomo N, et al. Recurrence and metastasis of lung cancer demonstrate decreased diffusion on diffusion-weighted magnetic resonance imaging. Asian Pac J Cancer Prev. 2014;15(16):6843–6848. doi:10.7314/apjcp.2014.15.16.6843
8. Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23(1):175–205. doi:10.1146/annurev.cellbio.23.090506.123406
9. Ritchie W. microRNA target prediction. Methods Mol Biol. 2017;1513:193–200. doi:10.1007/978-1-4939-6539-7_13
10. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
11. Liu Y, Li Y, Wang R, et al. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J Exp Clin Cancer Res. 2016;35(1):19. doi:10.1186/s13046-016-0296-0
12. Kong X, Zhang J, Li J, Shao J, Fang L. MiR-130a-3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell-like cells. Biochem Biophys Res Commun. 2018;501(2):486–493. doi:10.1016/j.bbrc.2018.05.018
13. Wang Y, Zhang X, Tang W, et al. miR-130a upregulates mTOR pathway by targeting TSC1 and is transactivated by NF-kappaB in high-grade serous ovarian carcinoma. Cell Death Differ. 2017;24(12):2089–2100. doi:10.1038/cdd.2017.129
14. Yang H, Zhang H, Ge S, et al. Exosome-derived miR-130a activates angiogenesis in gastric cancer by targeting C-MYB in vascular endothelial cells. Mol Ther. 2018;26(10):2466–2475. doi:10.1016/j.ymthe.2018.07.023
15. Zheng Y, Xiang L, Chen M, Xiang C. MicroRNA130a inhibits the proliferation, migration and invasive ability of hepatocellular carcinoma cells by downregulating rho-kinase 2. Mol Med Rep. 2018;18(3):3077–3084. doi:10.3892/mmr.2018.9283
16. Koul R, Rathod S, Dubey A, Bashir B, Chowdhury A. Comparison of 7th and 8th editions of the UICC/AJCC TNM staging for non-small cell lung cancer in a non-metastatic North American cohort undergoing primary radiation treatment. Lung Cancer. 2018;123:116–120. doi:10.1016/j.lungcan.2018.06.029
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
18. Verma V, Lautenschlaeger T. MicroRNAs in non-small cell lung cancer invasion and metastasis: from the perspective of the radiation oncologist. Expert Rev Anticancer Ther. 2016;16(7):767–774. doi:10.1080/14737140.2016.1191950
19. Lemjabbar-Alaoui H, Hassan OU, Yang YW, Buchanan P. Lung cancer: biology and treatment options. Biochim Biophys Acta. 2015;1856(2):189–210. doi:10.1016/j.bbcan.2015.08.002
20. Watanabe SI, Nakagawa K, Suzuki K, et al. Neoadjuvant and adjuvant therapy for stage III non-small cell lung cancer. Jpn J Clin Oncol. 2017;47(12):1112–1118. doi:10.1093/jjco/hyx147
21. Li WF, Dai H, Ou Q, Zuo GQ, Liu CA. Overexpression of microRNA-30a-5p inhibits liver cancer cell proliferation and induces apoptosis by targeting MTDH/PTEN/AKT pathway. Tumour Biol. 2016;37(5):5885–5895. doi:10.1007/s13277-015-4456-1
22. Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21(27):8284–8292. doi:10.3748/wjg.v21.i27.8284
23. Yu XF, Wang J, Ou N, et al. The role of miR-130a-3p and SPOCK1 in tobacco exposed bronchial epithelial BEAS-2B transformed cells: comparison to A549 and H1299 lung cancer cell lines. J Toxicol Environ Health A. 2019;82(15):862–869. doi:10.1080/15287394.2019.1664479
24. Lin L, Lin H, Wang L, Wang B, Hao X, Shi Y. miR-130a regulates macrophage polarization and is associated with non-small cell lung cancer. Oncol Rep. 2015;34(6):3088–3096. doi:10.3892/or.2015.4301
25. Liu B, Qu J, Xu F, et al. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6(11):9445–9456. doi:10.18632/oncotarget.3255
26. Fernandez S, Risolino M, Mandia N, et al. miR-340 inhibits tumor cell proliferation and induces apoptosis by targeting multiple negative regulators of p27 in non-small cell lung cancer. Oncogene. 2015;34(25):3240–3250. doi:10.1038/onc.2014.267
27. Yang RY, Hsu DK, Liu FT. Expression of galectin-3 modulates T-cell growth and apoptosis. Proc Natl Acad Sci U S A. 1996;93(13):6737–6742. doi:10.1073/pnas.93.13.6737
28. Sun W, Hu S, Zu Y, Deng Y. KLF3 is a crucial regulator of metastasis by controlling STAT3 expression in lung cancer. Mol Carcinog. 2019;58(11):1933–1945. doi:10.1002/mc.23072
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.