Back to Journals » Cancer Management and Research » Volume 13
miR-1298-5p Influences the Malignancy Phenotypes of Breast Cancer Cells by Inhibiting CXCL11
Received 28 August 2020
Accepted for publication 11 December 2020
Published 11 January 2021 Volume 2021:13 Pages 133—145
DOI https://doi.org/10.2147/CMAR.S279121
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Jie Zhang, Dawei Hu
Department of Breast Surgery, The Affiliated Hospital of Chengde Medical College, Chengde, Hebei 067000, People’s Republic of China
Correspondence: Dawei Hu
Department of Breast Surgery, The Affiliated Hospital of Chengde Medical College, No. 36 Nanyingzi Street, Chengde, Hebei 067000, People’s Republic of China
Tel +86 15633142697
Email [email protected]
Background: Breast cancer (BC) has deleterious effects on women’s health worldwide, yet its molecular mechanism remains unclear.
Objective: This study aimed to discover the underlying mechanism used by miR-1298-5p to regulate CXCL11 in BC.
Methods: Microarray analysis was performed to identify the key mRNA and miRNA involved in BC. The expression of miR-1298-5p and CXCL11 mRNA in BC clinical tissues and cell lines was detected using quantitative reverse transcription PCR (RT-qPCR), while the demonstration of intra- and extra-cellular CXCL11 protein was measured using western-blotting or ELISA assay. CCK-8, BrdU ELISA, colony formation, wound healing, and cell adhesion assays were carried out to determine cell viability, cell proliferation, colony formation, cell migration and adhesion phenotypes, respectively. A dual-luciferase assay kit was also employed to confirm the predicted binding scheme between miR-1298-5p and CXCL11.
Results: Microarray analysis confirmed miR-1298-5p and CXCL11 as the miRNA and mRNA to be further investigated in BC. After observing low-level miR-1298-5p and high-level CXCL11 in BC clinical tissues and cell lines, it was discovered that miR-1298-5p inhibited the phenotypes of BC cells, while CXCL11 promoted the tumorigenesis of BC cells. Findings indicated that miR-1298-5p attenuated the promotive effect of CXCL11 on BC cell phenotypes.
Conclusion: This research revealed that miR-1298-5p could influence the malignancy phenotypes of BC cells by inhibiting CXCL11.
Keywords: miR-1298-5p, breast cancer, CXCL11, phenotypes
Introduction
Breast cancer (BC) has been reported to affect the quality of life of women around the globe, especially those in developed countries. One study, for instance, reported that 3–10% of women showed distant metastases at initial presentation.1 However, in the last two decades, tremendous achievements have been made in surgery, adjuvant radio-chemotherapy, target therapy and other immunotherapies, and the 5-year survival rate of BC has been improved greatly, reaching as high as 90%.2–6 Despite these achievements, researchers believe that the identification of BC biomarkers will potentially upgrade BC therapies and offer more insights into the molecular mechanism of BC pathogenesis.
Regarded as a small single-stranded non-coding RNA with a length of about 22 nucleotides, microRNAs (miRNAs) can take a crucial role in protein-coding gene expression via multiple pathways, including RNA cleavage and translation inhibition. More specifically, the seed sequence of miRNAs can bind not only to 3 UTR but also to other parts of mRNAs (ie, introns, 5 UTR and exons of their target genes) to regulate the expression of target genes.7,8 This regulation can suppress the expression of the targeted genes at the post-transcriptional level.9 Besides, recent research indicated that miRNAs could play an irreplaceable role in cancerous growth. It was uncovered in one research that miR-1298-5p played an anti-tumor role in KRAS oncogene-induced tumor growth in vivo and in vitro 10 Moreover, miR-1298 was demonstrated to be downregulated in patients with non-small cell lung cancer, thereby prolonging their survival time.11 Although the investigation of the effect of miR-1298-5p on BC cells is still limited, it is hypothesized that miR-1298-5p might act as an anti-tumor miRNA in BC based on the previous studies that explored the impact of miR-1298-5p on other cancer types.
Apart from miRNAs, research has revealed that C-X-C Motif Chemokine Ligand 11 (CXCL11), a member of the CXC chemokine superfamily, could influence BC progression. As a member of this superfamily, CXCL11 has a small size of approximately 8 to 10 kDa.12 This molecular substance encodes the protein in activated T-cells to induce the chemotactic response, and it is regarded as the dominant ligand for CXCR3.12–14 In recent years, CXCL11 has been reported to be an oncogene in different cancers such as prostate cancer, colorectal cancer, and liver cancer.15–17 In their study, Liu et al found that CXCL11 was associated with the cytokine-cytokine receptor interaction pathway and that it might be a biomarker for assessing neoadjuvant chemotherapy in BC.18 However, scientists are yet to demystify the specific function of CXCL11 in BC.
This research aimed to investigate the underlying mechanism used by miR-1298-5p to regulate CXCL11 in BC. To identify miR-1298-5p and CXCL11 as the desired study objects in BC, microarray analysis of the GEO DataSets was performed. Several cellular experiments were also conducted to discern the interaction and function of miR-1298-5p and CXCL11 in BC cells. This study is relevant and worthwhile in that it has the potential to improve BC treatments.
Materials and Methods
Bioinformatics Analysis
GSE139038 from the GEO DataSets was the mRNA expression profile, which included 24 normal tissue samples and 41 BC samples. GSE143564 from the GEO database was the miRNA expression profile, which included three non-tumor breast tissue samples and three BC tissue samples. The upregulation of differentially expressed genes (DEGs) of BC from GSE139038 was chosen using the limma package with an adjusted P-value < 0.05 and a log-fold change (logFC) > 0. The downregulation of differentially expressed miRNAs of BC from GSE143564 was selected using the limma package with a P-value < 0.05 and a logFC < 0. GEPIA was utilized to show the expression of the key gene in the tumor and normal samples. The Kaplan Meier-Plotter (http://kmplot.com/analysis/) was subsequently employed to show the relationship between gene expression and BC prognosis. The starBase v2.0, an open-source platform, was used to predict the target relationship between miRNAs and mRNA.19 Venny 2.1.0 was also leveraged to identify the overlapping miRNAs.
Clinical Samples
A total of 50 paired frozen BC tumor tissues and adjacent non-tumor tissues were collected at the time of the surgery from the Affiliated Hospital of Chengde Medical College. The matched non-tumor breast tissues were collected from the same breast 2 mm away from the tumor. This study was approved by the Ethics Committee of the Affiliated Hospital of Chengde Medical College, and all patients were allowed to complete the written consent forms before the commencement of the study. The histopathology of the tissues was confirmed by three independent pathologists. The clinical characteristics of the patients with BC are shown in Table 1. Considering the heterogeneity of BC, the immunohistochemistry classification is significant. Thus, the expression data of miR-1298-5p and CXCL11 in ER+ and ER- patients were provided (Supplementary Figure 1).
 |
Table 1 Clinical Parameters of Patients with Breast Cancer in This Study |
Cell Culture
MCF10A, the human epithelial cell line, was purchased from ATCC (Cat#: CRL-10317, USA). The human BC cell lines purchased from ATCC (USA) included T-47D (Cat#: HTB-133), MDA-MB-231 (Cat#: HTB-26), SK-BR-3 (Cat#: HTB-30), MCF7 (Cat#: HTB-22), and BT-474 (Cat#: HTB-20). MCF10A and T-47D cells, SK-BR-3 cells, MDA-MB-231 and MCF7 cells, and BT-474 cells were cultured in an RPMI 1640 medium (Gibco, USA), a McCoy’s 5a medium (Cat#: 30–2007, ATCC, USA), a DMEM medium (Gibco, USA), and a Hybri-Care medium (Cat#: 46-X, ATCC, USA), respectively. FBS (10%), penicillin (1%), and streptomycin (1%) was added to all the media in an atmosphere containing 5% CO2 at 37°C.
miRNA and mRNA Detection
The total RNAs were isolated from the clinical samples and cells with the RNAprep Pure Cell/Bacteria Kit (Tiangen Biochemical, China). After performing RNA quantification at 260 nm, the RevertAid First Strand cDNA Synthesis Kit (Fermentas, USA) was used to reverse-transcribe 1 μg RNA into cDNA. With the SYBR Green PCR Master Mix (Applied Biosystems, USA), quantitative reverse transcription PCR (RT-qPCR) was subsequently performed to analyze the expression of miR-1298-5p and CXCL11. U6 and GAPDH were eventually used as the endogenous control for miR-1298-5p and CXCL11, respectively. The primer sequences are illustrated in Table 2.
 |
Table 2 The Sequences of the Primers in This Study |
Cell Transfection
Negative control plasmids, miR-1298-5p mimic, and CXCL11 overexpression vectors used in this study were designed by and purchased from GeneCopoeia (Guangzhou, China). The full-length sequences of CXCL11 were then amplified and cloned into pcDNA3.1 plasmid to construct CXCL11 overexpression vectors. Next, 1.5×104 T-47D and SK-BR-3 cells were seeded into the 6-well plates. After culturing the cells overnight, the miR-1298-5p mimic, CXCL11 overexpression vectors, or negative control plasmids were transfected into the cells using Lipofectamine 3000 Reagent (Cat#: L3000015, ThermoFisher, USA). After 40-hour transfection, the transfection efficiency was assessed using RT-qPCR.
Cell Viability Detection
The transfected T-47D and SK-BR-3 cells were seeded into 96-well plates at a concentration of 1500 cells per well. After performing cell transfection, the Cell Counting Kit-8 (CCK-8) was utilized to evaluate cell viability on Day 1, Day 2, Day 3, and Day 4. After adding 10 μL CCK8 solution into every well, the cells were cultured in an incubator for another 4 hours at 37°C. Finally, the optical density at 450 nm was read using a microplate reader.
Cell Proliferation Detection
Cell proliferation was detected using the BrdU ELISA assay. Similar to the CCK-8 assay, the transfected cells (1500 cells/well) were seeded into 96-well plates. After culturing the cells for 4 days in an incubator, 10× BrdU solution (10 μL/well) was added to the cells for an incubation period of 4 hours. After washing the cells with PBS three times, 100 μL diluted anti-BrdU antibody was added to each well, and the mixture was incubated for 1 hour. Following that, 100 μL of the diluted secondary antibody was added to each well for an incubation period of 1 hour. Finally, the absorbance of each well was read at 450 nm with a microplate reader.
Colony Formation Detection
The colony formation assay was performed to evaluate the malignancy growth of the cells. The transfected T-47D and SK-BR-3 cells were first seeded into 6-well plates at a concentration of 1×105 per well and then incubated at 37°C for two weeks. In each cell well, crystal violet staining was then carried out to visualize the colonies. Finally, the number of colonies was counted to demonstrate the growth of T-47D and SK-BR-3 cells.
Cell Migration Detection
The migration capability of the BC cells was assessed with a wound-healing assay system. First, 3.5×105/well T-47D and SK-BR-3 cells were seeded into 12-well plates. The cells were subsequently cultured until the cell confluence was more than 90%. Next, 200 μL micropipette tips were used to scratch the cell monolayers. After the exfoliated cells were washed away with PBS, the cell monolayers were photographed under an inverted microscope. The cells were subsequently cultured in a serum-free medium for 24 hours. The cell monolayer was then photographed again under an inverted microscope.
Cell Adhesion Detection
The transfected T-47D and SK-BR-3 suspended by the serum-free medium were placed in the 96-well plate (10000 cells/well) coated with 10 μg/mL laminin or collagen. After blocking the nonspecific binding with 1% BSA, the cells were allowed to adhere to the plate at 37°C for 30 and 60 min. Next, the adherent cells were fixed with 4% paraformaldehyde, and the non-adherent cells were washed with PBS. Subsequently, the fixed cells were stained with 2.5% crystal violet in methanol. The absorbance was eventually measured at 450 nm using a microplate reader.
Dual-Luciferase Assay
Based on the starBase prediction on the binding site between CXCL11 and miR-1298-5p, the CXCL11 3ʹUTR binding sequences were mutated. The wild-type CXCL11 3ʹUTR (CXCL11 3ʹUTR-WT) and mutant CXCL11 3ʹUTR (CXCL11 3ʹUTR-Mut) were inserted into the pGL3-basic vector. The two BC cell lines (T-47D and SK-BR-3) were then transiently co-transfected with miR-1298-5p mimic, the negative control, pGL3-CXCL11-3′UTR-WT, and pGL3-CXCL11-3′UTR-Mut. After 24-hour transfection, the luciferase activities were measured using the Secrete-Pair Dual Luminescence Assay Kit (GeneCopoeia, USA).
Western Blotting Assay
The total protein was extracted from the cells using the RIPA buffer, which contained 5mM EDTA and PMSF. After determining the protein concentration with the BCA protein assay kit (Pierce, USA), 30 μg protein were separated with 12% SDS-PAGE gels and then transferred onto the PVDF membranes (Millipore, MA, USA). The membranes were then blocked in 5% skim milk and incubated with the CXCL11 antibody (Cat#: ab9955, Abcam, UK), GAPDH (Cat#: ab9485, Abcam, UK), and anti-rabbit secondary antibodies. The protein was eventually measured using the ECL kit (Pierce, USA).
CXCL11 Detection in the Supernatants of Culture Media
The concentrations of CXCL11 in the supernatants of T-47D and SK-BR-3 cells’ culture media were measured using the Human CXCL11/I-TAC Quantikine ELISA Kit (R&D Systems, USA). The measurement was done according to the manufacturer’s protocol. Subsequently, 100 μL/well of Assay Diluent RD1-68 was added to each well of the 96-well microplate coated with a monoclonal antibody. The culture media (100 μL) collected were supplemented, and the plate was incubated for 2 hours at room temperature. After washing the cells three times with the wash buffer, 200 μL of Human CXCL11 Conjugate was added to each well, and the mixture was incubated for another 2 hours at room temperature. The cells were then washed three times again. Afterward, 200 μL of Substrate Solution was added to the well, and the mixture was incubated at room temperature in the dark for 30 minutes. Finally, 50 μL Stop Solution was supplemented, and the absorbance of each well was read at 450 nm using a microplate reader. The concentration of CXCL11 in the supernatant was calculated using the standard curve.
Statistical Analysis
GraphPad Prism 7 was employed to perform statistical analysis. Three independent experiments were conducted, and all the data were presented in the form of mean ± SD. The 2-tailed Student’s t-test was applied to analyze the differences between the two groups, whereas the one-way analysis of variance (ANOVA) was utilized to evaluate the differences between more than two groups. Moreover, the associations between miR-1298-5p and CXCL11 expression and clinicopathological features were analyzed using the chi-square test. P-value < 0.05 was considered to be statistically significant.
Result
mRNA and miRNA Identification in BC
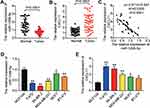
GSE139038 and GSE143564 were used to identify the key mRNA and miRNA in BC, respectively. A total of 1322 upregulated DEGs in BC samples were identified, with the adjacent P-value < 0.05 and the logFC > 0. The top 10 upregulated DEGs are depicted in Figure 1A. The second most upregulated DEG of interest was CXCL11, and GEPIA analysis revealed that CXCL11 expression was significantly elevated in 1085 BC samples compared with the 291 normal samples (Figure 1B). The results of Kaplan Meier-plotter analysis indicated that high-level CXCL11 exhibited poor survival in BC (Figure 1C). Hence, CXCL11 was selected as the gene of interest that required further investigations in BC. A growing number of studies have demonstrated that CXCL11 plays an oncogenic role in a variety of cancers, including prostate cancer, colorectal cancer and liver cancer,15–17 a result that further highlights the potential value of CXCL11 in the pathogenesis of BC.
 |
Figure 1 CXCL11 and miR-1298-5p were identified as our interested mRNA and miRNA. (A) The top 10 most upregulated DEGs from GSE139038 data analysis are shown. GSE139038 is the gene microarray profile of BC. Limma was performed to screen out the DEGs with an adj. (adjusted) P value smaller than 0.05 and a logFC (fold change) bigger than 0. DEGs: differentially expressed genes. (B) The expression of CXCL11 in 1085 tumor and 291 normal breast samples was assessed by GEPIA. BRCA, breast invasive carcinoma. *P<0.01. (C) The effect of CXCL11 on survival of BC patients was analyzed by Kaplan Meier Plotter (http://kmplot.com). (D) The overlapping miRNAs between the dataset of the miRNAs that target CXCL11 predicted by starBase (http://starbase.sysu.edu.cn/) and the dataset of downregulated DEGs from GSE143564 analysis. GSE143564 is the miRNA microarray profile of BC. Limma was applied to analyze the downregulated miRNAs in BC with P < 0.05 and logFC < 0. |
After using starBase to predict the miRNAs associated with CXCL11, it was found that a total of 8 miRNAs could bind to CXCL11. Besides, after performing limma analysis with a P-value < 0.05 and a logFC < 0, a total of 2111 miRNAs were identified to be downregulated in BC. Finally, hsa-miR-219a-5p and hsa-miR-1298-5p were discovered to be the overlapping miRNAs in target miRNAs of CXCL11, and they downregulated miRNAs from GSE143564 (Figure 1D).
miR-1298-5p and CXCL11 Expression in BC
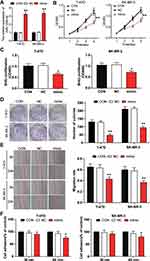
To verify the effect of miR-1298-5p and CXCL11 on BC, RT-qPCR was first used to detect the expression of miR-1298-5p and CXCL11 in BC tissues and non-tumor tissues. It was observed that the expression of miR-1298-5p decreased by 53.3% in the tumor tissues (Figure 2A), while the expression of CXCL11 increased by 2.25-fold in the tumor tissues (Figure 2B). The results of Pearson correlation analysis indicated the negative correlation of miR-1298-5p and CXCL11 in BC tissues (Figure 2C). Besides, miR-1298-5p with low expression and CXCL11 with high expression were observed in 5 BC cell lines compared with the normal cell line (Figure 2D and E). Because the difference in the expression of miR-1298-5p and CXCL11 in T-47D and SK-BR-3 was the largest, T-47D and SK-BR-3 cell lines were selected for subsequent cellular experiments.
Biological Functions of miR-1298-5p in BC Cells
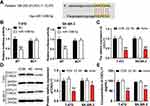
Several experiments were conducted to further investigate the function of miR-1298-5p in BC. To upregulate miR-1298-5p, miR-1298-5p mimic was transfected into the two BC cell lines, T-47D and SK-BR-3. The result showed that the expression of miR-1298-5p grew by more than 3-fold in BC cells (Figure 3A). The CCK-8 assay result also revealed that the viability of T-47D and SK-BR-3 cells in the miR-1298-5p overexpression group significantly decreased on Day 3 and Day 4 compared with the control group (Figure 3B). After performing the BrdU ELISA assay, it was noticed that the absorbance of the miR-1298-5p mimic group declined by 38% in T-47D cells and fell by 31% in SK-BR-3 cells compared with the control group, a result that indicated that miR-1298-5p mimic could suppress cell proliferation (Figure 3C). Moreover, the colony-formation assay result revealed a 60% decrease in the colony number of T-47D and SK-BR-3 cells with the transfection of miR-1298-5p mimic in contrast to the control group (Figure 3D). As for the outcome of the wound healing assay, a 30% decrease in the migration rate was observed in these two BC cell lines after transfecting miR-1298-5p mimic (Figure 3E). What’s more, the cell adhesion capability shrank by approximately 20% in T-47D and SK-BR-3 cells after transfecting miR-1298-5p mimic for 60 min (Figure 3F). Taken together, these findings indicated that miR-1298-5p inhibited the malignancy phenotypes of BC cells.
miR-1298-5p Targeted CXCL11 in BC
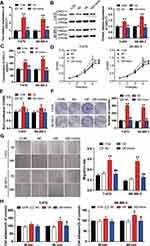
The blinding site of miR-1298-5p in the 3ʹ-UTR of CXCL11 was predicted using starBase. The outcome suggested that CXCL11 might be a potential target of miR-1298-5p (Figure 4A). Besides, the result of the dual-luciferase assays showed that the luciferase activity contracted by almost 50% when the T-47D and SK-BR-3 cells were co-transfected with miR-1298-5p mimic and wild-type CXCL11 3ʹUTR. However, the co-transfection of miR-1298-5p mimic and mutant CXCL11 3ʹUTR had no effect on the luciferase activity in T-47D and SK-BR-3 cells (Figure 4B). Moreover, the expression of CXCL11 mRNA decreased by more than 50% when the T-47D and SK-BR-3 cells were transfected with miR-1298-5p mimic (Figure 4C). The result of the Western blotting assay further proved that the expression of CXCL11 protein declined in the miR-1298-5p group compared with the control group; the expression of CXCL11 protein showed a 50% decrease in T-47D cells (Figure 4D). The CXCL11 ELISA assay result demonstrated that the protein level of CXCL11, which was secreted into the culture medium, were also significantly downregulated (around 50%) in the miR-1298-5p group compared with the control group (Figure 4E). Collectively, these data not only validated the binding scheme predicted by starBase between CXCL11 mRNA 3ʹUTR and miR-1298-5p but also showed that the expression of CXCL11 could be inhibited by miR-1298-5p.
Relation Between CXCL11 and miR-1298-5p in BC
To verify whether miR-1298-5p could regulate the biological function of CXCL11 in BC, the CXCL11 overexpression vectors and miR-1298-5p mimic were first transfected or co-transfected into BC cells. The expression of CXCL11 was detected using RT-qPCR, and findings showed that the expression of CXCL11 in the CXCL11 overexpression group increased by more than 2.5-fold compared with the control group in two BC cell lines. Also, the co-transfection of CXCL11 overexpression vectors and miR-1298-5p mimic resulted in a 2.5-fold decrease in CXCL11 expression compared with the CXCL11 overexpression group (Figure 5A). Similarly, the outcome of the Western blotting assay showed that in contrast to the control group, the protein expression of CXCL11 increased, respectively, by 1.44-fold and 1.85-fold in T-47D and SK-BR-3 cells with CXCL11 overexpression; however, the co-transfection of miR-1298-5p mimic and CXCL11 overexpression plasmid reversed the change (Figure 5B). Moreover, the result of the CXCL11 ELISA assay showed a significant increase in the protein level of CXCL11 in the supernatant of culture media for T-47D and SK-BR-3 cells with CXCL11 overexpression, while the co-transfection of miR-1298-5p mimic and CXCL11 overexpression plasmid effectively reversed the effect caused by CXCL11 overexpression (Figure 5C). Experimental results later indicated that the BC cells transfected with CXCL11 overexpression vectors exhibited stronger cell viability compared to the control group. However, it was found that the BC cells co-transfected with CXCL11 overexpression vectors and miR-1298-5p mimic indicated weaker cell viability compared to the CXCL11 overexpression group (Figure 5D). Similar to the outcome of the CCK-8 assay, the result of the BrdU ELISA assay indicated that the cell proliferation capability could be enhanced by upregulating CXCL11. Nevertheless, the co-transfection of CXCL11 overexpression vectors and miR-1298-5p mimic could alleviate the cell-proliferation promotive function of CXCL11 (Figure 5E). Besides, the colony formation assay result revealed that the colony number of T-47D and SK-BR-3 cells in the CXCL11 overexpression group was upregulated by about 3 fold compared to the control group. Nonetheless, this consequence was completely reversed by co-transfecting CXCL11 overexpression vectors and miR-1298-5p mimic (Figure 5F). After performing the wound healing assay, it was found that CXCL11 overexpression resulted in a 2-fold increase in the migration rate in T-47D and a 3-fold increase in the migration rate in SK-BR-3. The migration rate in the co-transfection group, however, was significantly reduced compared to the CXCL11 overexpression group (Figure 5G). Moreover, the result of the cell adhesion assay showed the same trend as that of the wound healing assay. This similarity suggested that CXCL11 overexpression promoted cell adhesion; nonetheless, its promotive function could be reversed by transfecting miR-1298-5p mimic (Figure 5H). Overall, these findings demonstrated that CXCL11 promoted the malignancy phenotypes of BC cells even though miR-1298-5p could mitigate this enhancement function.
Discussion
In this research, we first conducted a microarray analysis to identify miR-1298-5p and CXCL11 as our miRNA and mRNA of interest in BC. Our findings revealed that miR-1298-5p was downregulated in BC tissues and cells, while CXCL11 was upregulated in BC tissues and cells. We noticed that miR-1298-5p overexpression inhibited the viability, proliferation, colony formation, migration, and adhesion abilities of BC cells. Unlike miR-1298-5p, CXCL11 promoted the malignancy phenotypes of BC cells by enhancing cell viability, cell proliferation, colony formation, cell migration, and cell adhesion. We also discovered that miR-1298-5p inhibited CXCL11 expression in BC cells by binding to CXCL11 mRNA 3ʹUTR to alleviate its function.
Nonetheless, the literature has limited information regarding the tumor-suppressive role of miR-1298. More specifically, miR-1298-5p was proved to be downregulated in bladder cancer clinical tissues, and its upregulation inhibited the phenotypes of bladder cancer cells by suppressing Cx43 expression.20 Zhou et al, for instance, found that miR-1298 impeded the tumor growth induced by the oncogene KRAS in vivo and in vitro.10 Another research confirmed the downregulation of miR-1298 in non-small cell lung cancer and its suppressive role in cancer phenotypes.11 In this study, miR-1298-5p was significantly downregulated in BC tissue samples, and the upregulation of miR-1298-5p significantly impaired cell phenotypes such as the viability, proliferation, colony formation, migration, and adhesion abilities of BC cells. For this reason, we speculated that miR-1298-5p could suppress BC growth.
Furthermore, researchers have uncovered that CXCL11 could influence the development of colorectal cancer, BC, gastric cancer, and esophageal cancer.21–23 After collecting colorectal cancer tissue samples, Gao et al found that CXCL11 was overexpressed in colorectal cancer tissues and that CXCL11 silence reduced tumor growth and metastasis in vivo.16 Liu et al, on the other hand, used the digital gene expression sequencing technique to compare the gene expression profiling between the pathological complete response samples and the non-pathological complete response samples. They discovered that CXCL11 was upregulated in pathological complete response samples associated with the cytokine-cytokine receptor interaction pathway.18 In our study, we performed microarray analysis and found that CXCL11 was the second most significantly upregulated gene in BC. The microarray analysis results were also validated in our collected tissue samples. We conducted several functional experiments and proved that CXCL11 contributed to the malignancy phenotypes of BC cells in vitro. Our finding was consistent with the result of the previous BC studies in terms of the ability of CXCL11 to promote tumor cell migration and invasion phenotypes.22,24,25 Besides, we found that miR-1298-5p was the upstream miRNA of CXCL11 and that it could inhibit CXCL11 expression by binding to CXCL11 3ʹUTR, a result that was different from that of the previous study.
Furthermore, chemokines represent a complex net system, and they can influence the tumor microenvironment in synergy with other regulators.26 Despite our significant findings in this study, we do not have sufficient evidence to conclude that CXCL11 chemokine is responsible for the entire BC phenotype. This is because many other chemokines have been reported to be involved in BC. For instance, it was found that chemokine C-C motif ligand 5 (CCL5) interacted with interleukin-1β (IL-1β), tumor necrosis factor-α (TNF-α), and chemokine CCL2 to promote BC progression.27,28 Chemokines also played an essential function in communication signaling among various immune cells, such as influencing cancer progression directly.29 For instance, a clinical-stage monoclonal antibody that neutralized IL-8, HuMax-IL8 was shown to significantly decrease the recruitment of polymorphonuclear myeloid-derived suppressor cells at the tumor site and to enhance the immune-mediated killing.30 In another experiment, IL-1β-deficient mice showed low-level CCL2 and CSF-1, thus hampering the recruitment of monocytes and inhibiting the ability of monocytes to transform into macrophages. Therefore, the immune cells could not infiltrate into tumors to suppress BC progression.31 Many other studies have also revealed that chemokines work together32,33 and interact with the immune system34,35 to regulate BC progression. These studies highlight the joint action between the chemokines and the immune system in BC malignancy development. Even though our study elucidated the role of CXCL11 chemokine in BC, further investigation is needed regarding its interaction with immune systems and other chemokines.
Clinical trials regarding the utility of chemokines in the BC treatments have been performed. A decade ago, a clinical trial revealed that the overexpression of C-X-C motif chemokine receptor 4 (CXCR4) could predict poor outcomes for patients with locally advanced BC who underwent neoadjuvant chemotherapy.36 Reparixin, an investigational allosteric inhibitor of CXCR1/2, was demonstrated in 2017 to be effective and safe for HER-2-negative metastatic BC patients when it was used with paclitaxel weekly.37 In addition, a high CXCL13 level was associated with favorable distant disease-free survival outcomes for patients with early BC, especially those with triple-negative BC.38 In a recent clinical trial, an oral CCL2 inhibitor-propagermanium (PG) was used as an anti-metastatic drug for BC patients, thus demonstrating a manageable safety profile.39 These clinical trials indicated that chemokines/chemokine receptors/chemokine ligands could be promising prognostic markers and anti-BC targets in BC. Besides, we found in our analysis that modifying CXCL11 expression could affect BC cell phenotypes in vitro. In short, this finding indicated its potential as an anti-BC target that could be used in clinical practice.
Conclusion
In summary, this study demonstrated that miR-1298-5p could directly regulate the expression of CXCL11. Put simply, miR-1298-5p was found to contribute to the malignancy phenotypes of BC cells by suppressing CXCL11. This effect might provide a new theoretical basis for BC treatments. Nevertheless, the influence of miR-1298-5p and CXCL11 in vivo needs to be further explored in future research.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
The present study was approved by the Ethics Committee of Affiliated Hospital of Chengde Medical College (Chengde, China). All patients signed written informed consent.
Consent for Publication
Consent for publication was obtained from the participants.
Funding
Funding information is not available.
Disclosure
The authors declare that they have no conflict of interests.
References
1. Desille-Gbaguidi H, Avigdor S, Body G, Ouldamer L. Survival impact of primary site surgery on metastatic breast cancer patients at diagnosis. J Gynecol Obstet Hum Reprod. 2019;48(3):171–177. doi:10.1016/j.jogoh.2018.10.014
2. Im SA, Lu YS, Bardia A, et al. Overall Survival with Ribociclib plus Endocrine Therapy in Breast Cancer. N Engl J Med. 2019;381(4):307–316. doi:10.1056/NEJMoa1903765
3. Kuerer HM, Newman LA, Smith TL, et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol. 1999;17(2):460–469. doi:10.1200/JCO.1999.17.2.460
4. Ahmad A. Breast Cancer Statistics: recent Trends. Adv Exp Med Biol. 2019;1152:1–7.
5. Hendrick RE, Baker JA, Helvie MA. Breast cancer deaths averted over 3 decades. Cancer. 2019;125(9):1482–1488.
6. Guanghui R, Xiaoyan H, Shuyi Y, Jun C, Guobin Q. An efficient or methodical review of immunotherapy against breast cancer. J Biochem Mol Toxicol. 2019;33(8):e22339.
7. Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab. 2018;15:68.
8. Wilczynska A, Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22(1):22–33.
9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
10. Zhou Y, Dang J, Chang KY, et al. miR-1298 Inhibits Mutant KRAS-Driven Tumor Growth by Repressing FAK and LAMB3. Cancer Res. 2016;76(19):5777–5787. doi:10.1158/0008-5472.CAN-15-2936
11. Du Z, Wu J, Wang J, et al. MicroRNA-1298 is downregulated in non-small cell lung cancer and suppresses tumor progression in tumor cells. Diagn Pathol. 2019;14(1):132. doi:10.1186/s13000-019-0911-4
12. Rotondi M, Chiovato L, Romagnani S, Serio M, Romagnani P. Role of chemokines in endocrine autoimmune diseases. Endocr Rev. 2007;28(5):492–520. doi:10.1210/er.2006-0044
13. Schroepf S, Kappler R, Brand S, et al. Strong overexpression of CXCR3 axis components in childhood inflammatory bowel disease. Inflamm Bowel Dis. 2010;16(11):1882–1890. doi:10.1002/ibd.21312
14. Lee EY, Lee ZH, Song YW. CXCL10 and autoimmune diseases. Autoimmun Rev. 2009;8(5):379–383. doi:10.1016/j.autrev.2008.12.002
15. Wang Y, Xu H, Si L, et al. MiR-206 inhibits proliferation and migration of prostate cancer cells by targeting CXCL11. Prostate. 2018;78(7):479–490. doi:10.1002/pros.23468
16. Gao YJ, Liu L, Li S, et al. Down-regulation of CXCL11 inhibits colorectal cancer cell growth and epithelial-mesenchymal transition. Onco Targets Ther. 2018;11:7333–7343. doi:10.2147/OTT.S167872
17. Wei X, You X, Zhang J, Zhou C. MicroRNA-1305 Inhibits the Stemness of LCSCs and Tumorigenesis by Repressing the UBE2T-Dependent Akt-Signaling Pathway. Mol Ther Nucleic Acids. 2019;16:721–732. doi:10.1016/j.omtn.2019.04.013
18. Liu X, Jin G, Qian J, et al. Digital gene expression profiling analysis and its application in the identification of genes associated with improved response to neoadjuvant chemotherapy in breast cancer. World J Surg Oncol. 2018;16(1):82. doi:10.1186/s12957-018-1380-z
19. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi:10.1093/nar/gkt1248
20. Li G, Sun L, Mu Z, et al. MicroRNA-1298-5p inhibits cell proliferation and invasion of bladder cancer via downregulating connexin 43. Biochem Cell Biol. 2019.
21. Zhang Y, Zhao W, Li S, et al. CXCL11 promotes self-renewal and tumorigenicity of alpha2delta1(+) liver tumor-initiating cells through CXCR3/ERK1/2 signaling. Cancer Lett. 2019;449:163–171. doi:10.1016/j.canlet.2019.02.016
22. Barrow-McGee R, Procter J, Owen J, et al. Real-time ex vivo perfusion of human lymph nodes invaded by cancer (REPLICANT): a feasibility study. J Pathol. 2019:2.
23. Camargo MC, Song M, Shimazu T, et al. Circulating Inflammation Markers and Risk of Gastric and Esophageal Cancers: A Case-Cohort Study Within the Japan Public Health Center-Based Prospective Study. Cancer Epidemiol Biomarkers Prev. 2019;28(4):829–832. doi:10.1158/1055-9965.EPI-18-1157
24. Kundu N, Ma X, Brox R, et al. The Chemokine Receptor CXCR3 Isoform B Drives Breast Cancer Stem Cells. Breast Cancer. 2019;13:1178223419873628. doi:10.1177/1178223419873628
25. Bauer D, Mazzio E, Soliman KFA. Whole Transcriptomic Analysis of Apigenin on TNFalpha Immuno-activated MDA-MB-231 Breast Cancer Cells. Cancer Genomics Proteomics. 2019;16(6):421–431. doi:10.21873/cgp.20146
26. Soria G, Ben-Baruch A. The inflammatory chemokines CCL2 and CCL5 in breast cancer. Cancer Lett. 2008;267(2):271–285. doi:10.1016/j.canlet.2008.03.018
27. Soria G, Yaal-Hahoshen N, Azenshtein E, et al. Concomitant expression of the chemokines RANTES and MCP-1 in human breast cancer: a basis for tumor-promoting interactions. Cytokine. 2008;44(1):191–200. doi:10.1016/j.cyto.2008.08.002
28. Soria G, Ofri-Shahak M, Haas I, et al. Inflammatory mediators in breast cancer: coordinated expression of TNFalpha & IL-1beta with CCL2 & CCL5 and effects on epithelial-to-mesenchymal transition. BMC Cancer. 2011;11(1):130. doi:10.1186/1471-2407-11-130
29. Chow MT, Luster AD. Chemokines in cancer. Cancer Immunol Res. 2014;2(12):1125–1131. doi:10.1158/2326-6066.CIR-14-0160
30. Dominguez C, McCampbell KK, David JM, Palena C. Neutralization of IL-8 decreases tumor PMN-MDSCs and reduces mesenchymalization of claudin-low triple-negative breast cancer. JCI Insight. 2017;2(21):21. doi:10.1172/jci.insight.94296
31. Kaplanov I, Carmi Y, Kornetsky R, et al. Blocking IL-1β reverses the immunosuppression in mouse breast cancer and synergizes with anti–PD-1 for tumor abrogation. Proc Natl Acad Sci U S A. 2019;116(4):1361–1369. doi:10.1073/pnas.1812266115
32. Brummer G, Acevedo DS, Hu Q, et al. Chemokine Signaling Facilitates Early-Stage Breast Cancer Survival and Invasion through Fibroblast-Dependent Mechanisms. Mol Cancer Res. 2018;16(2):296–308. doi:10.1158/1541-7786.MCR-17-0308
33. Kitamura T, Pollard JW. Therapeutic potential of chemokine signal inhibition for metastatic breast cancer. Pharmacological Research. 2015;100:266–270. doi:10.1016/j.phrs.2015.08.004
34. Svensson S, Abrahamsson A, Rodriguez GV, et al. CCL2 and CCL5 Are Novel Therapeutic Targets for Estrogen-Dependent Breast Cancer. Clin Cancer Res. 2015;21(16):3794–3805. doi:10.1158/1078-0432.CCR-15-0204
35. Bedognetti D, Wang E, Marincola FM. Meta-analysis and metagenes: CXCL-13-driven signature as a robust marker of intratumoral immune response and predictor of breast cancer chemotherapeutic outcome. Oncoimmunology. 2014;3(4):e28727. doi:10.4161/onci.28727
36. Holm NT, Abreo F, Johnson LW, Li BDL, Chu QD. Elevated chemokine receptor CXCR4 expression in primary tumors following neoadjuvant chemotherapy predicts poor outcomes for patients with locally advanced breast cancer (LABC). Breast Cancer Res Treat. 2009;113(2):293–299. doi:10.1007/s10549-008-9921-8
37. Schott AF, Goldstein LJ, Cristofanilli M, et al. Phase Ib Pilot Study to Evaluate Reparixin in Combination with Weekly Paclitaxel in Patients with HER-2–Negative Metastatic Breast Cancer. Clin Cancer Res. 2017;2(21):5358–5365. doi:10.1158/1078-0432.CCR-16-2748
38. Schmidt M, Weyer-Elberich V, Hengstler JG, et al. Prognostic impact of CD4-positive T cell subsets in early breast cancer: a study based on the FinHer trial patient population. Breast Cancer Res. 2018;20(1):15.
39. Masuda T, Noda M, Kogawa T, et al. Phase I dose-escalation trial to repurpose propagermanium, an oral CCL2 inhibitor, in patients with breast cancer. Cancer Sci. 2020;111(3):924–931.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.