Back to Journals » OncoTargets and Therapy » Volume 7
miR-129 as a novel therapeutic target and biomarker in gastrointestinal cancer
Authors Fesler A, Zhai H, Ju J
Received 5 April 2014
Accepted for publication 15 May 2014
Published 21 August 2014 Volume 2014:7 Pages 1481—1485
DOI https://doi.org/10.2147/OTT.S65548
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Andrew Fesler, Haiyan Zhai, Jingfang Ju
Department of Pathology, Stony Brook Medicine, Stony Brook University, Stony Brook, NY, USA
Abstract: In the last decade, cancer research efforts in the field of noncoding microRNA (miRNA) have been growing exponentially. miRNA-based therapeutics have been tested in both preclinical and clinical settings, and miRNA-based cancer diagnostics and prognostics have moved into clinics to help better manage cancer treatment. A growing body of evidence in recent literature suggests miRNA-129 plays important roles in gastrointestinal cancer, including gastric, colorectal, hepatocellular carcinoma, and esophageal cancer. In this review, we focus on accumulating evidence demonstrating the key roles that miRNA-129 plays in tumorigenesis, disease progression, chemoresistance, proliferation, and cell cycle control. Understanding the emerging roles and mechanisms of miRNA-129 in cancer will help us realize the therapeutic and diagnostic/prognostic potential.
Keywords: miR-129, cancer, therapeutics, biomarker, 5-FU
Introduction
With only 2%–3% of RNA in the cell represented by protein coding messenger RNA, there is great interest and importance in understanding noncoding RNA (eg, ribosomal RNA, transfer RNA, microRNA [miRNA], and long noncoding RNAs), which makes up the vast majority of cellular RNA. The functions of noncoding RNA, and in particular miRNAs, have been studied extensively for their roles in fundamental biology and human diseases, including cancer.
miRNAs are a class of noncoding, 20–22-nucleotide RNAs that have been clearly demonstrated to carry out crucial regulatory functions in the cell. miRNAs are processed from larger pre-miRNAs by the RNase III enzyme Dicer (DICER1) into miRNA duplexes.1 One strand of this duplex associates with the RNA-induced silencing complex, whereas the other strand is generally degraded by cellular nucleases.1 The miRNA–RNA-induced silencing complex binds to specific mRNA targets via imperfect base pairing in the 3′-untranslated region, leading to translational repression or degradation of these mRNAs.2–4 Beyond this function, miRNA has also been shown to target messenger RNA in the coding region, as well as the 5′-untranslated region.5,6 In addition, miRNA can also regulate transcription through targeting of promoter regions.7,8 The importance of understanding these molecules has been recognized through the awarding of the Nobel Prize in Physiology or Medicine to Andrew Fire and Craig Mello for their work on RNA interference.9 Although it was shown in 1993 that lin-4, a small noncoding RNA in Caenorhabditis elegans, could influence development, it was not till 2002 that it was revealed that miRNA can, in fact, play a relevant role in cancer.2,10,11 This sparked great interest in elucidating the dynamic roles miRNA may play in the development and progression of cancer and led to the rapid expansion of this field.
To fully understand gene expression changes in cancer, it is essential to know not only the levels of individual mRNAs but also the extent to which mRNAs are translated into their corresponding proteins and the miRNAs that regulate these processes. Both genetic and epigenetic alterations affect the expression of miRNAs in cancer. Mounting evidence suggests miRNAs can function as either oncogenes or tumor-suppressor genes. As a result, some of them will have potential as therapeutic targets and/or biomarkers. As work has been done investigating the roles of miRNA in cancer, some specific miRNAs have shown great promise. One particular miRNA in which there has been growing interest because of its targeting of important genes associated with apoptosis, cell cycle, and chemoresistance is miR-129, and evidence is beginning to show that this miRNA may play an important role in cancer.
Deregulated miR-129 expression
miR-129 was initially reported to be downregulated in undifferentiated gastric cancer tissue when compared with miR-129 expression in the paired normal controls.12 miR-129 expression was found to be reduced in colorectal cancer,13,14 gastric cancer,15 and liver cancer.16 In esophageal cancer, there have been conflicting reports concerning miR-129 expression, with some work indicating it is reduced in cancer tissue compared with normal tissue17,18 and others claiming it is increased in tumor tissue.19 The reduced expression of miR-129 is significantly associated with disease stage of colorectal cancer and progression.14 Such reduction of miR-129 in colorectal cancer was a result of hypermethylation of the miR-129 promoter.13 Similar results were found in gastric cancer, where hypermethylation was associated with miR-129 reduction in gastric cancer cell lines, and reduced expression was associated with poor clinical-pathological features.15,20 This epigenetic regulation of miR-129 has also been demonstrated by different groups in hepatocellular carcinoma (HCC).21,22 Thus, there is evidence from multiple different types of cancer that miR-129 is reduced in cancer compared with normal tissue, and this deregulation is likely carried out by hypermethylation. This suggests that miR-129 may play a role as a tumor suppressor.
Functional significance of miR-129
The functional significance of miR-129 in cancer has been investigated in detail in recent years. The first evidence that one of the key targets affected by miR-129 is SOX4 was reported in endometrial cancer. miR-129 expression was reduced by epigenetic repression, which leads to the upregulation of SOX4 in endometrial cancer.23 Similar results have been found in esophageal cancer and HCC.18,22 SOX4 is a potent transcription factor linked to several key pathways for oncogenesis, such as transforming growth factor β, Hedgehog, and Notch signaling. Such mechanisms have also been validated to be true in gastric cancer.24 It appears that epigenetic silencing of miR-129 in gastric cancer is a key event leading to the upregulation of SOX4 during tumorigenesis. SOX4 is overexpressed in several types of cancer and may play an important role in metastasis and tumor progression.25 As it is well known that a single miRNA can influence translation of hundreds of mRNA target transcripts, in several types of cancer, including gastric cancer, miR-129 was also found to regulate cell cycle and target CDK6.20,26
In addition to SOX4 and CDK6, recent studies have also shown miR-129 to be a key regulator of apoptosis by suppressing the expression of Bcl2 in colon cancer.14 This targeting of Bcl2 may be of critical significance in colon cancer, as Bcl2 is expressed in colon tumor tissue but not in normal tissue and may play an important role in the development of the disease.27 miR-129 also affects cell cycle by suppressing the expression of E2F3. More importantly, miR-129 overexpression restores chemosensitivity to 5-fluorouracil (5-FU) treatment in colon cancer by suppressing 5-FU therapeutic target, thymidylate synthase, and Bcl2.14 These results suggest that miR-129 has great potential as a key tumor suppressor in colorectal cancer, liver cancer, and gastric cancer by affecting cell proliferation, cell cycle, tumorigenesis, and chemosensitivity through multiple targets and pathways. The current understanding of miR-129, with its critical targets, pathways, and cellular functions, is illustrated in Figure 1. miR-129 targets that have been identified in gastrointestinal cancers are summarized in Table 1.
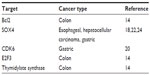  | Table 1 Targets of microRNA-129 in gastrointestinal cancers |
Therapeutic potential of miR-129 in gastrointestinal cancer
As we have discussed earlier, miR-129 affects several key targets and pathways in cancer. Thus, it is reasonable to assume that approaches to restore miR-129 expression will likely be beneficial to cancer treatment. This notion is also based on the fact that miR-129 is epigenetically silenced in gastric and colorectal cancer. It is logical to reason that restoration of miR-129 expression will have tumor-suppressive potential, either alone or in combination with chemotherapy for developing novel therapeutic strategies for treating advanced colorectal cancer, liver cancer, and gastric cancer. Our recent studies demonstrated that restoration of miR-129 in vivo can sensitize colon cancer tumor xenografts to 5-FU treatment.14 Further investigation will be necessary to validate the potential of miR-129 as a therapeutic, as well as considerations concerning effective delivery and modifications to enhance activity.
Potential of miR-129 as a diagnostic, predictive, and prognostic biomarker
There has been growing interest in the potential of miRNAs as cancer biomarkers. Identification of biomarkers has been a significant challenge, and those that are used, such as carcinoembryonic antigen for colon cancer, often demonstrate low sensitivity. mRNA-based biomarkers are difficult in clinical settings because of issues related to mRNA degradation and the stringent requirement of sample quality. miRNAs represent a unique opportunity for biomarker research, as they have increased stability in formalin fixed paraffin embedded (FFPE) compared with mRNA biomarkers and also are stable extracellularly and may be useful as circulating biomarkers.28–30 miRNA function is also tissue-specific, potentially increasing the value of miRNAs as tissue-specific biomarkers. Many miRNAs have shown potential to be used as biomarkers thus far.31 miR-129 is among these potential miRNA biomarkers, and it has been clearly demonstrated that the loss of miR-129 expression is significantly associated with disease progression in colorectal cancer, gastric cancer, and liver carcinoma. As a result, miR-129 has the potential to be used as a companion diagnostic biomarker, along with clinical histopathological diagnosis. In addition to the clinical relevance of miR-129 in gastric tumor tissues, the expression level of miR-129 is also reported to be a potential noninvasive approach for gastric cancer diagnosis, using gastric juice.32 The reduced expression of miR-129 has been shown in colorectal cancer via epigenetic silencing by promoter methylation. We further revealed a progressive loss of miR-129 in normal colon, colon adenoma, and stage I–IV colorectal cancer samples. The expression levels of miR-129 were significantly reduced in stage III and nearly completely missing in stage IV colorectal cancer patient samples.14 miR-129 expression is closely associated with response to 5-FU-based chemotherapy and patient survival.
Conclusion and perspectives
miR-129 has drawn much attention from the research community. It targets several mRNAs that play important roles in tumor development and progression, thus giving it great potential as a novel therapeutic molecule for treating cancer. However, we must bear in mind that the function of miR-129 is disease- and context-specific, and a thorough understanding of these different functions would be crucial for further steps aimed at developing a miR-129-based therapeutic. With miR-34 currently in clinical trials, all eyes are on the progress of miRNA-based therapeutics. Although many hurdles (eg, effective delivery of miRNA) may lie ahead, we are optimistic that the future of miRNA-based therapeutics will be bright and that miR-129 may have the potential to be next in line for therapeutic development.
Acknowledgments
The work was supported by grant funding from the National Institutes of Health (R01CA155019 and R33CA147966 to JJ).
Disclosure
The authors report no conflicts of interest in this work.
References
Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. | |
Lee RC, Feinbaum RL, Ambros V. The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. | |
Pillai RS, Bhattacharyya SN, Artus CG, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309(5740):1573–1576. | |
Ruvkun G. Clarifications on miRNA and cancer. Science. 2006; 311(5757):36–37. | |
Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–9672. | |
Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. | |
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–1613. | |
Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18(3):223–231. | |
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. | |
Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C elegans. Cell. 1993;75(5):855–862. | |
Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. | |
Katada T, Ishiguro H, Kuwabara Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34(2):537–542. | |
Bandres E, Agirre X, Bitarte N, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–2743. | |
Karaayvaz M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis. 2013;4(6):e659. | |
Tsai KW, Wu CW, Hu LY, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129(11):2600–2610. | |
Liu Y, Hei Y, Shu Q, et al. VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS ONE. 2012;7(4):e35800. | |
Chen X, Hu H, Guan X, et al. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. Int J Cancer. 2012;130(7):1607–1613. | |
Kang M, Li Y, Liu W, et al. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32(1):51–58. | |
Ogawa R, Ishiguro H, Kuwabara Y, et al. Expression profiling of micro-RNAs in human esophageal squamous cell carcinoma using RT-PCR. Med Mol Morphol. 2009;42(2):102–109. | |
Yu X, Song H, Xia T, et al. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532(1):87–93. | |
Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer. 2013;52(7):636–643. | |
Chen X, Zhang L, Zhang T, et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2013;33(3):476–486. | |
Huang YW, Liu JC, Deatherage DE, et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69(23):9038–9046. | |
Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394(4):1047–1052. | |
Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32(29):3397–3409. | |
Wu J, Qian J, Li C, et al. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9(9):1809–1818. | |
Sun N, Meng Q, Tian A. Expressions of the anti-apoptotic genes Bag-1 and Bcl-2 in colon cancer and their relationship. Am J Surg. 2010;200(3):341–345. | |
Xi Y, Nakajima G, Gavin E, et al. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–1674. | |
Cho WC. Circulating MicroRNAs as Minimally Invasive Biomarkers for Cancer Theragnosis and Prognosis. Front Genet. 2011;2:7. | |
Fesler A, Jiang J, Zhai H, Ju J. Circulating microRNA testing for the early diagnosis and follow-up of colorectal cancer patients. Mol Diagn Ther. 2014;18(3):303–308. | |
Cho WC. MicroRNAs: potential biomarkers for cancer diagnosis, prognosis and targets for therapy. Int J Biochem Cell Biol. 2010;42(8):1273–1281. | |
Yu X, Luo L, Wu Y, et al. Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30(1):365. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.