Back to Journals » Cancer Management and Research » Volume 13
MiR-129-5p Suppresses Cell Proliferation of Human Osteosarcoma Cancer by Down-Regulating LncRNA Lnc712
Authors Yu Y, Zuo W, Cai W, Xu Y, Liu W, Zhao Z
Received 14 October 2020
Accepted for publication 14 February 2021
Published 10 March 2021 Volume 2021:13 Pages 2259—2264
DOI https://doi.org/10.2147/CMAR.S284078
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Yifan Yu, Wenshan Zuo, Wei Cai, Yong Xu, Weidong Liu, Zexue Zhao
Department of Orthopaedics, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, Huai’an City, Nanjing Province, 223300, People’s Republic of China
Correspondence: Wenshan Zuo
Department of Orthopaedics, The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University, No. 1 Huanghe West Road, Huaiyin District, Huai’an City, Nanjing Province, 223300, People’s Republic of China
Email [email protected]
Introduction: Lnc712 has been characterized as an oncogenic lncRNA in breast cancer. This study aimed to investigate the role of Lnc712 in osteosarcoma (OS).
Methods: OS and paired non-tumor tissues were collected from 58 OS patients. Expression of Lnc712 and miR-129-5p in paired tissue samples was determined by RT-qPCR. Lnc712 and miR-129-5p expression was achieved in OS cells to study the interaction between them. Cell proliferation was analyzed by CCK-8 assay.
Results: Lnc712 was upregulated in OS and was inversely correlated with miR-129-5p. In OS cells, Lnc712 overexpression failed to significantly affect miR-129-5p, while miR-129-5p overexpression led to downregulated Lnc712. Cell proliferation showed that Lnc712 overexpression resulted in increased cell proliferation rate. MiR-129-5p overexpression played an opposite role and reversed the effect of Lnc712 overexpression.
Discussion: MiR-129-5p may suppress cell proliferation of OS by down-regulating Lnc712.
Keywords: Lnc712, osteosarcoma, miR-129-5p, proliferation
Introduction
Osteosarcoma (OS) as a common type of bone cancer mostly originates from osteoblast cells mainly affects teenagers and adults younger than 30 years.1 OS is rare type of malignancy with an annual incidence rate of 3–4 out of one million people in the United States.2 Although OS is rarely curable, with proper treatment, such as ablative surgery, more than 70% of patients with regional OS can survive longer than 5 years.3 However, metastasis to other parts of the human body, such as other bones, brain and lungs, is common in OS patients.4,5 Once distant metastasis occurred, the 5-year overall survival rate will decrease to below 30%.6 Therefore, novel therapeutic approaches are still needed to further improve the survival of OS patients.
The exact cause is unknown.7 However, previous studies on the molecular pathogenesis of OS have revealed that nearly all aspects of the development and progression of OS require the involvement of molecular players.8,9 With the increased understanding of the functionality of these molecular players, targeted therapy, which aims to suppress cancer progression by regulating related gene expression, has come out.10 Non-coding RNAs (ncRNAs), such as the short miRNAs and long lncRNAs, are not involved in protein-coding but regulate gene expression at multiple levels to regulate human diseases.11,12 In the development and progression of OS, lncRNAs and miRNAs regulate multiple cell behaviors, such as proliferation, migration and invasion, to participate in cancer biology.13,14 Therefore, ncRNAs are promising targets for the development of targeted therapy. However, the function of most ncRNAs remains unclear. Lnc712 has been characterized as an oncogenic lncRNA in breast cancer,15 while its role in OS is unknown. We predicted that Lnc712 may be targeted by miR-129-5p, which is also a critical player in cancer biology.16 This study aimed to investigate the role of Lnc712 in OS and to explore its interaction with miR-129-5p.
Patients and Methods
OS Patients
From March 2017 to March 2020, we enrolled 58 OS patients (36 males and 22 females; 11 to 30 years; 20.4 ± 4.1 years) at The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University. Ethics Committee of this hospital approved this study before the enrollment of patients. No recurrent patients were included and all the 58 OS patients were newly diagnosed cases. In view of the fact that other clinical disorders or therapies may also affect the expression of genes that we were working on, patients complicated with other clinical disorders or patients received any therapies for any clinical disorders within 3 months before this study were excluded. All patients or their parents signed informed consent. Procedures operated in this research were completed in keeping with the standards set out in the Announcement of Helsinki and laboratory guidelines of research in China.
OS Tissues and Cells
Fine needle aspiration was performed all 58 OS patients to collect paired OS and non-tumor tissues. Following confirmation by histopathological exam, all tissue samples were immediately subjected to total RNA extractions and subsequent experiments.
MG-63 and Hs 3.T human OS cell lines (ATCC, USA) were used. Cells were cultivated in medium composed of 90% RPMI 1640 and 10% FBS. Cells were cultivated at 37°C in a 5% CO2 incubator (95%) humidity. For subcultivation, a ratio of 1:8 was used. Cells were harvested at about confluence of 85% to perform subsequent experiments.
Vectors, miRNAs and Cell Transfection
Backbone vector expressing Lnc712 was constructed with pcDNA3.1 vector (Invitrogen). Mimic of miR-129-5p and negative control (NC) miRNA were also from Invitrogen. Vector (1μg) or miRNA (50 nM) was transfected into 108 cells through Lipofectamine® 2000 (Invitrogen)-mediated transient transfections. To perform NC experiment, cells were transfected with either NC miRNA or empty vector. To perform control (C) experiment, cells without transfections were cultivated under normal conditions until the end of experiments. Incubation with transfection mixture was performed for 6h, followed by washing with fresh medium. Cells were cultivated for 48h after transfections prior to subsequent experiments.
RNA Preparation
RNAs were isolated from paired non-tumor and OS tissues, as well as MG-63 and Hs 3.T cells using Ribozol reagent (Invitrogen). To perform genomic DNA removal, DNase I (Invitrogen) was used to incubate with all RNA samples for 2h at 37°C. RNAs were separated by 5% Urine-PAGE gel to check RNA integrity. RNA purity was checked by determining the ratio of OD 260/280.
RT-qPCR
RNA samples with a ratio of OD 260/280 close to 2.0, which indicates pure RNA samples, were subjected to reverse transcriptions using a Reverse Transcription System (A2790, Promega Corporation) to synthesize cDNA samples. With cDNA samples as template, SYBR Green Master Mix (Bio-Rad) was used to prepare qPCRs with 18S rRNA as internal control to measure the levels of Lnc712 expression.
To determine the expression of mature miR-129-5p, poly(A) addition to mature miRNAs, reverse transcriptions and qPCRs were performed with U6 as internal control. All the three steps were performed using All-in-One™ miRNA qRT-PCR Reagent Kit (Genecopoeia). Ct values of three technical replicates included in each experiment were normalized to internal controls using 2−∆∆CT method. Sequences of primers used were: 5´-AAATACCTCACCCTCATCTATACCAAC-3´ (forward) and 5´-TTTCCCGTTGCCATTGAT-3´ (reverse) for Lnc712; 5´-CTACCACATCCAAGGAAGCA-3´ (forward) and 5´-TTTTTCGTCACTACCTCCCCG-3´ (reverse) for 18S rRNA. Forward primer of miR-129-5p was 5´-CTTTTTGCGGTCTGGGCT-3´. Reverse primer of miR-129-5p and U6 primers were included in the kit.
Cell Proliferation Assay
Following transfections, MG-63 and Hs 3.T cells were subjected to cell proliferation assays using Cell Counting Kit 8 (CCK8, ab228554, Abcam). Cells were transferred to each well of a 96-well cell culture plate with 4000 cells in 0.1 mL medium per well. At 37°C, cells were cultivated and OD values (450 nm) were measured every 24h for a total of 96h. At 2h before the measurement of OD values, CCK-8 solution was added to 10%. OD values were normalized to C group at 96h (100%) to plot proliferation curves. This assay was performed in at least three biological replicates, in each biological replicate three technical replicates were included.
Statistical Analysis
Gene expression levels in paired tissues measured by RT-qPCR were expressed as average values. Mean±SD value was used to express data of three independent replicates of other experiments. Paired t-test was used for the exploration of differences between paired t-test. ANOVA Tukey’s test was used to explore significant differences among more than 2 groups. Correlation analysis was performed by linear regression. P<0.05 was statistically significant.
Results
Altered Expression of Lnc712 and MiR-129-5p Was Observed in OS
Lnc712 and miR-129-5p expression in paired non-tumor and OS tissues collected from OS patients (n=58) was determined by RT-qPCR. Compared to non-tumor tissues, OS tissues exhibited significantly higher levels of Lnc712 expression (Figure 1A, p<0.001). In contrast, OS tissues exhibited significantly lower levels of miR-129-5p expression in comparison to non-tumor tissues (Figure 1B, p<0.001). Therefore, altered expression of Lnc712 and miR-129-5p may participate in OS.
Lnc712 and MiR-129-5p Were Inversely Correlated
Correlation analysis showed that Lnc712 and miR-129-5p were significantly and inversely correlated across OS tissues (Figure 2A). In addition, a reverse and significant correlation between Lnc712 and miR-129-5p was also observed across non-tumor tissues (Figure 2B). Therefore, Lnc712 and miR-129-5p may interact with each other.
 |
Figure 2 Lnc712 and miR-129-5p were inversely correlated. Linear regression was performed to analyze the correlations between Lnc712 and miR-129-5p across OS tissues (A) and non-tumor tissues (B). |
MiR-129-5p May Target Lnc712 to Downregulate It in OS Tissues
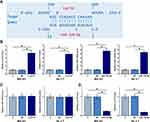
The potential interaction between Lnc712 and miR-129-5p was predicted by IntaRNA 2.0 (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp). It was observed that miR-129-5p may target Lnc712 (Figure 3A). To explore the interaction between Lnc712 and miR-129-5p, Lnc712 expression vector or miR-129-5p mimic was transfected into MG-63 and Hs 3.T cells. Transfections were confirmed by RT-qPCR (Figure 3B, p<0.05). It was observed that cells transfected with Lnc712 expression vector showed no significant alterations in the expression levels of miR-129-5p (Figure 3C). In contrast, cells with miR-129-5p mimic transfection showed significantly lower levels of Lnc712 expression (Figure 3D, p<0.05).
MiR-129-5p Targeted Lnc712 to Suppress Cell Proliferation
The roles of Lnc712 and miR-129-5p in regulating the proliferation of MG-63 and Hs 3.T cells were explored by cell proliferation assay. It was observed that Lnc712 overexpression resulted in increased cell proliferation rate. MiR-129-5p overexpression played an opposite role and reversed the effect of Lnc712 overexpression (Figure 4, p<0.05).
Discussion
This study analyzed the interactions between miR-129-5p and Lnc712 in OS. We found that miR-129-5p was downregulated in OS and Lnc712 was upregulated in OS. In addition, miR-129-5p may target Lnc712 to suppress the proliferation of OS cells.
In a recent study, Cui et al reported that Lnc712 was overexpressed in breast cancer and it may interact with heat-shock protein 90 to mediate the binding of heat-shock protein 90 to cell division cycle 37, thereby promoting cancer cell division and proliferation.15 In a recent study, Lnc712 was found to be significantly overexpressed in hepatocellular carcinoma and predicted poor survival of patients.16 In addition, Lnc712 may interact with miR-142-3p to upregulate Bach-1, thereby promoting cancer cell proliferation and metastasis.16 Based on our knowledge, the involvement of Lnc712 in other types of cancers remains unclear. Specifically, the upstream regulator of this lncRNA in cancer biology is unknown. In this study, we first reported the upregulation of Lnc712 in OS. In addition, overexpression of Lnc712 resulted in the increased proliferation of OS cells. OS is a type of sarcoma, which has very different mechanism from breast cancer. Our study and the previous study15 showed that Lnc712 could promote the proliferation of cancer cells in two different types of cancers. Therefore, we may speculate that Lnc712 may also promote cancer cell proliferation in other type of cancers. More studies are needed to elucidate the functionality of Lnc712 in other cancers.
MiR-129-5p has been reported to play tumor-suppressive role in several types of cancers, such as OS.18,19 It is known that miR-129-5p gene is methylated in OS and demethylation of miR-129-5p suppresses the malignant phenotype of OS cells.18 In another study, miR-129-5p was reported to target DLK1 to reduce the stemness of OS cells.19 Based on our knowledges, all previous studies reported the targeting of protein-coding genes by miR-129-5p. In this study, we reported that miR-129-5p could also target a lncRNA in OS cells. In addition, we observed that miR-129-5p and Lnc712 were inversely correlated across both non-tumor and OS tissues. Therefore, the targeting of Lnc712 by miR-129-5p may exist under both pathological and physiological conditions. Future studies may focus on the role of the interaction between Lnc712 and miR-129-5p in physiological processes.
However, this study has several limitations. First, due to the low incidence of OS, this study only analyzed a small number of patients. Moreover, the in vivo interaction between miR-129-5p and Lnc712 was unknown. Future studies are needed to include more patients and perform in vivo animal model experiments to further confirm our conclusions.
In conclusion, miR-129-5p is downregulated in OS and Lnc712 is upregulated in OS. MiR-129-5p can target Lnc712 to suppress the proliferation of OS cells. The increased understating of the interaction between miR-129-5p and Lnc712 may provide novel target for the therapeutic treatment of OS.
Availability of Supporting Data
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the privacy of research participants.
Ethical Approval and Consent to Participate
All patients or their parents signed informed consent. All producers were approved by The Affiliated Huaian No.1 People’s Hospital of Nanjing Medical University Ethics Committee. This study was conducted in accordance with the Declaration of Helsinki.
Disclosure
The authors have no conflicts of interest to declare.
References
1. Klein MJ, Siegal GP. Osteosarcoma: anatomic and histologic variants. Am J Clin Pathol. 2006;125(4):555–581. doi:10.1309/UC6KQHLD9LV2KENN
2. Durfee RA, Mohammed M, Luu HH. Review of osteosarcoma and current management. Rheumatol Ther. 2016;3(2):221–243. doi:10.1007/s40744-016-0046-y
3. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi:10.1016/j.ocl.2015.08.022
4. Kovar H. Selective enhancer changes in osteosarcoma lung metastasis. Nat Med. 2018;24(2):126–127. doi:10.1038/nm.4487
5. Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16(5):543–556. doi:10.1586/14737140.2016.1168697
6. Song K, Song J, Lin K, et al. Survival analysis of patients with metastatic osteosarcoma: a surveillance, epidemiology, and end results population-based study. Int Orthop. 2019;43(8):1983–1991. doi:10.1007/s00264-019-04348-4
7. Kushlinskii NE, Fridman MV, Braga EA. Molecular mechanisms and microRNAs in osteosarcoma pathogenesis. Biochem (Mosc). 2016;81(4):315–328. doi:10.1134/S0006297916040027
8. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737–755. doi:10.1016/j.molmed.2017.06.004
9. Denduluri SK, Wang Z, Yan Z, et al. Molecular pathogenesis and therapeutic strategies of human osteosarcoma. J Biomed Res. 2015;30:30. doi:10.7555/JBR.29.20150075
10. Sayles LC, Breese MR, Koehne AL, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63. doi:10.1158/2159-8290.CD-17-1152
11. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15(1):38. doi:10.1186/s12935-015-0185-1
12. Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859(1):169–176. doi:10.1016/j.bbagrm.2015.06.015
13. Chen R, Wang G, Zheng Y, et al. Long non-coding RNAs in osteosarcoma. Oncotarget. 2017;8(12):20462–20475. doi:10.18632/oncotarget.14726
14. Han C, Wang W. MicroRNA-129-5p suppresses cell proliferation, migration and invasion via targeting ROCK1 in osteosarcoma. Mol Med Rep. 2018;17(3):4777–4784. doi:10.3892/mmr.2018.8374
15. Cui Y, Lu C, Zhang Z, et al. A long non-coding RNA Lnc712 regulates breast cancer cell proliferation. Int J Biol Sci. 2020;16(1):162–171. doi:10.7150/ijbs.36429
16. Cui D, Ni C. LncRNA Lnc712 promotes tumorigenesis in hepatocellular carcinoma by targeting miR-142-3p/Bach-1 axis. Cancer Manag Res. 2020;12:11285. doi:10.2147/CMAR.S254950
17. Ma L, Chen X, Li C, et al. miR-129-5p and-3p co-target WWP1 to suppress gastric cancer proliferation and migration. J Cell Biochem. 2019;120(5):7527–7538. doi:10.1002/jcb.28027
18. Long XH, Zhou YF, Peng AF, et al. Demethylation-mediated miR-129-5p up-regulation inhibits malignant phenotype of osteogenic osteosarcoma by targeting homo sapiens valosin-containing protein (VCP). Tumour Biol. 2015;36(5):3799–3806. doi:10.1007/s13277-014-3021-7
19. Zhang R, Tang T, Yu HM, et al. LncRNA DLX6-AS1/miR-129-5p/DLK1 axis aggravates stemness of osteosarcoma through Wnt signaling. Biochem Biophys Res Commun. 2018;507(1–4):260–266. doi:10.1016/j.bbrc.2018.11.019
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.