Back to Journals » Cancer Management and Research » Volume 10
miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA
Authors Yang X, Qiu J, Kang H, Wang Y, Qian J
Received 9 January 2018
Accepted for publication 3 May 2018
Published 16 November 2018 Volume 2018:10 Pages 5839—5853
DOI https://doi.org/10.2147/CMAR.S161990
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Dr Leylah Drusbosky
Xuehua Yang, Jianwei Qiu, Haifeng Kang, Yaming Wang, Junbo Qian
Department of Gastroenterology, The Second Affiliated Hospital of Nantong University, Nantong, Jiangsu, China
Background: MiR-125a-5p has been reported to be involved in the development and progression of various cancers. However, the biological function and underlying mechanisms in colorectal cancer(CRC) still remain unclear. Here, we explored the potential biological roles of miR-125a-5p in CRC.
Methods: The expression of miR-125a-5p was detected using quantitative real-time PCR (qRT-PCR), biological functions of miR-125a-5p were assessed by cell counting kit-8, wound-healing, transwell invasion, and human umbilical vein endothelial cell (HUVEC) tube formation assays in vitro and animal experiments in vivo.
Results: We found that miR-125a-5p was downregulated in CRC tissues and cell lines, it inhibited CRC cell proliferation, migration, and invasion and reduced the ability of HUVECs to form tubes. Moreover, we verifed that miR-125a-5p suppressed CRC growth and metastasis in vivo. Additionally, we showed that VEGFA, a direct target gene of miR-125a-5p, could reverse the inhibitory effect caused by miR-125a-5p overexpression.
Conclusion: miR-125a-5p might serve as a tumor suppressor in CRC and could be regarded as a potential therapeutic candidate for CRC.
Keywords: miR-125a-5p, colorectal cancer, VEGFA, overall survival
Introduction
Colorectal cancer (CRC) is the third most common and the fourth leading cause of tumor-related death globally.1 Despite the improvement in the chemotherapy and radiotherapy, the overall survival (OS) rate of CRC patients do not seem to be elevated substantially,2 and the underlying mechanism of the initiation and progression of CRC is largely unknown. Therefore, identifying the molecular mechanism of CRC is urgently needed.
MicroRNAs (miRNAs) are a group of small noncoding RNAs (18-25 nucleotides in length) that regulate their target gene expression via binding to the 3′untranslated regions (3′UTRs) of the corresponding target mRNA.3 Increasing evidences have confirmed that aberrantly expressed miRNAs played key roles in multiple biological processes of CRC, including proliferation, metastasis, apoptosis, drug resistance, and angiogenesis.4-7
miR-125a-5p, located on 19q13.41, has been reported to be aberrantly expressed and functioned as a tumor suppressor in various cancers, such as glioblastoma,8 hepatocelluar carcinoma,9 gastric cancer,10, 11 and nonsmall-cell lung cancer.12 However, the biological functions of miR-125a-5p in CRC are largely unknown.
In our study, we found that miR-125a-5p was downregulated in CRC cells and tissues, and the expression of which was markedly reversely correlated with OS. Moreover, we identified VEGFA might be a direct target gene of miR-125a-5p in CRC. Additionally, we also showed that miR-125a-5p inhibited tumor growth and metastasis in vitro and vivo. Therefore, we could conclude that miR-125a-5p acts as a tumor suppressor in CRC.
Materials and methods
Clinical tissues
Forty CRC tissues and matched adjacent normal tissues (ANTs) were collected from The Second Affiliated Hospital of Nantong University from January 2006 to December 2008. The tissues were immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction. None of these patients received radiotherapy or chemotherapy before surgical resection. Written informed consent was obtained from all these patients, and this study was approved by the Institutional Review Board of The Second Affiliated Hospital of Nantong University. Clinicopathological characteristics of these patients are listed in Table 1.
  | Table 1 Correlation between miR-125a-5p expression and different clinical characteristics Abbreviation: CEA, carcinoembryonic antigen. |
Cell culture
Human CRC cell lines HCT116, HT29, HCT8, SW620 and a normal colonic epithelial cell line (FHC) were obtained from American Type Culture Collection. These cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) with 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 10% fetal bovine serum (FBS; Thermo Fisher Scientific) at 37°C and supplied with 5% CO2 atmosphere.
Cell transfection
AgomiR-125a-5p, antagomiR-125a-5p, and corresponding negative control (agomiR-NC, antagomiR-125a-5p) were purchased from GenePharma (Shanghai, China). pcDNA3.1-VEGFA and blank vector were purchased from Genecreat (Wuhan, China). Small interference RNA (siVEGFA) was chemically synthesized (Applied Biologic Materials, Inc, Richmond, BC, Canada), and the sequence is listed in Table S1. HCT116 and HT29 cells were seeded into six-well plates prior to antagomiR-125a-5p (30 nM), agomiR-125a-5p (30 nM), pcDNA3.1-VEGFA (50 nM), siVEGFA (50 nM), or their negative control transfection, and transfetion was performed using Lipofectamine 2000 (Thermo Fisher Scientific) following the manufacturer’s instructions. The transfected cells were collected at 48 h after transfection.
Cell proliferation assay
The proliferation of cells was assessed using cell counting kit-8 assay (CCK-8; KeyGen BioTech, Nanjing, China) in accordance with the manufacturer’s instructions. Absorbance was measured at 450 nm using a microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Cell migration assay
A wound healing assay was performed to evaluate cell migration. Briefly, transfected cells were seeded into six-well plates (3×105 cells/well) and grown to 80%-90% confluence. Cells were scratched with 200 µL pipette tips and then washed with PBS. Cells were further cultured with a medium containing 2% FBS for 24 h. Images were captured by a microscope (Nikon Corporation, Tokyo, Japan) at 40× magnification.
Cell invasion assay
Cell invasion assay was performed using a 24-well transwell chamber with 8 μM pores (Corning Incorporated, Corning, NY, USA). Briefly, transfected cells were seeded into the upper transwell chambers precoated with Matrigel (BD Biosciences, San Jose, CA, USA). The lower chambers were loaded with 500 µL of DMEM containing 20% FBS. After 24 h, noninvading cells were removed by a cotton swab and invaded cells were fixed with 100% methanol, and then stained with 0.1% crystal violet. The number of invaded cells was counted under an inverted microscope (Nikon Corporation) at 200× magnification at five randomly selected fields.
Tube formation assay
HUVECs (4×105) were suspended in a mixture of tumor-conditioned medium (TCM, 300 µL) and DMEM containing 10% FBS (300 µL), and then seeded into a 96-well plate precoated with Matrigel (100 µL per well; BD Biosciences). Tube formation was observed after 6 h of incubation at 37°C and was imaged with an inverted microscope (Nikon Corporation).
Dual-luciferase reporter assay
The potential miR-125a-5p-binding sites in VEGFA-3′UTR were predicted by miRanda (www.microRNA.org). The wild-type (WT) VEGFA-3′UTR (WT-VEGFA-3′UTR) and mutant VEGFA-3′UTR (Mut-VEGFA-3′ UTR) containing the putative binding sites for miR-125a-5p were chemically synthesized and cloned into pmirGLO vector. The cells (293T and HCT116) were seeded into 24-well plates for 24 h and then cotranfected with pmirGLO-Mut-VEGFA-3′UTR or pmirGLO-WT-VEGFA-3′ UTR and agomiR-125-5p or agomiR-NC. Dual-luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA) to detect the Renilla and firefly luciferase activities after 24 h of transfection. Renilla luciferase activity was used for normalization.
Western blot assay
Protein was extracted from the cells by using a lysis buffer containing protease inhibitor (KeyGen BioTech) and was quantified with a bicinchoninic acid kit (KeyGen BioTech). Thirty micrograms of protein extracts were separated using 10% sodium dodecyl sulphate - polyacrylamide gel electrophoresis (SDS-PAGE) gel before transferring to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking with 5% nonfat milk for 1 h, the membranes were then incubated with primary antibodies at 4°C overnight: anti-VEGFA (1:1,000; Proteintech, Chicago, IL, USA), GAPDH (1:5,000; Proteintech), anti-VEGFR (1:1,000; Cell Signaling Technology, Beverly, MA, USA), anti-p-VEGFR2 (1:1,000; Cell Signaling Technology), and anti-p-Akt (1:1,000; Cell Signaling Technology). Next day, the membranes were incubated with secondary antibody for 1 h at room temperature. The protein bands were detected using an enhanced chemiluminescence reagent kit (KeyGen BioTech).
RNA isolation and quantitative RT-PCR
TRIzol reagent (Thermo Fisher Scientific) was used to extract total RNA in accordance with manufacturer’s instructions. miR-125a-5p expression was detected by a Hairpin-itTM microRNA and U6 snRNA normalization RT-PCR quantitation kit (GenePharma). For VEGFA mRNA, complementary DNA (cDNA) was synthesized using PrimeScriptTM RT reagent kit with gDNA eraser (Takara, Tokyo, Japan), and qRT-PCR was analyzed using SYBR Premix Ex Taq kit (Takara). The PCR primes for VEGFA and GAPDH are as follows: VEGFA: forward: 5′-TGGCTCACTGGCTTGCTCTA-3′, reverse: 5′-ATCCAACTGCACCGTCACAG-3′; GAPDH: forward: 5′-GGTGGTCTCCTCTGACTTCAA-3′, reverse: 5′-GTTGCTGTAGCCAAATTCGTTGT-3′. The relative expression of miRNA or mRNA was analyzed using 2-ΔΔCT method. All results were normalized to GAPDH or U6.
Immunohistochemistry
The CRC tissues and ANTs were collected, paraffin embedded, and cut into 4-µm-thick sections. Sections were incubated with rabbit anti-VEGFA antibodies at 4°C overnight, followed by incubation with secondary antibodies. The sections were visualized under an inverted microscope (Nikon Corporation) at 200× or 400× magnification. The intensity of staining was scored by two independent pathologists in the following four categories: no staining=0, weak staining=1, moderate staining=2, and strong staining=3. The stain-positive samples were scored into four grades: 0 (0%), 1 (1-33%), 2 (34%-66%), and 3 (67%-100%). The final immunohistochemistry score was calculated by multiplying the percentage of positive cells with the intensity score.
In vivo experiments
All in vivo experiments were approved by the Animal Care Committee of The Second Affiliated Hospital of Nantong University and were performed in accordance with the animal experimental guidelines of NanTong University. For tumor xenograft mode, 1×107 HCT116cells were subcutaneously injected into the flank of 5-week-old male BABL/c nude mice that were randomly divided into two groups (n=6 in each group). After tumor formation, 1.5 nmol of agimiR-125a-5p or agomiR-NC was injected into the tumors. The injections were performed seven times (days 6, 8, 10, 12, 14, 16, 18). After 2 days of last injection, the mice were sacrificed, and the volume of tumors was measured. The volume of tumor was calculated with the formula: 0.5×length×width2.
For metastasis experiments, 1×106 HCT116 cells in 0.1 mL PBS were injected into the tail vein in two groups of mice. The mice were sacrificed after 7 weeks of injection, and lung tissues were isolated from the mice and tissues sections were stained with H&E. The numbers of metastatic nodules were counted using an inverted microscope (Nikon Corporation) at 100× and 200× magnifications.
Statistical analysis
All experiments were performed in triplicate, and statistical analysis was performed using SPSS 19.0 software (Chicago, IL, USA). Differences between the two groups were evaluated using Student’s t-test (two-tailed), and correlations between VEGFA and miR-125a-5p were analyzed by Spearman rank correlation. Kaplan-Meier method was applied to assess the OS. The survival curves were compared with log-rank test. Follow-up time was censored if the patient was lost to follow-up. Cox proportional hazards model was used to perform multivariate analysis and calculate the 95% CI. Results of experiments are presented as mean±SD. P<0.05 was considered to indicate statistical significance.
Results
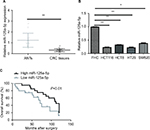
miR-125a-5p was downregulated in CRC tissues and cell lines
qRT-PCR was used to detect the miR-125a-5p expression levels in 40 pairs of CRC tissues and matched ANTs. miR-125a-5p was markedly decreased in CRC tissues compared with ANTs (Figure 1A). Additionally, low levels of miR-125a-5p were also detected in HCT116, HCT8, HT29, and SW620 cells compared with that in FHC cells (Figure 1B), and HCT116 and HT29 cells were selected for further studies in the following experiments.
Next, we investigated the association of miR-125a-5p expression with several clinical parameters. CRC patients were divided into two groups (showing low and high miR-125a-5p expression) with median miR-125a-5p expression levels regarded as the cutoff point between the two groups. We found that miR-125a-5p expression levels in CRC were obviously associated with TNM stage. However, the expression of miR-125a-5p was not significantly associated with gender, age, tumor location, lymph node, differentiation, tumor size, and carcinoembryonic antigen (CEA) (Table 1).
Kaplan-Meier survival analyses indicated that CRC patients with low miR-125a-5p expression had a significantly poorer OS than those with high miR-125a-5p expression (Figure 1C). Moreover, multivariate cox regression analysis showed that miR-125a-5p could be an independent prognostic indicator for OS (Table 2).
  | Table 2 Univariate and multivariate analyses for OS in patients with CRC Abbreviations: OS, overall survival; CRC, colorectal cancer; CEA, carcinoembryonic antigen. |
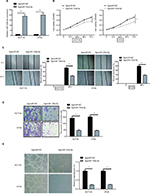
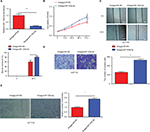
miR-125a-5p inhibited CRC cell proliferation, migration, invasion, and angiogenesis in vitro
To assess the biological function of miR-125a-5p in the initiation and progression of CRC, HCT116 and HT29 cells were transfected with agomiR-125a-5p or agomiR-NC. The efficiency of transfection was detected using qRT-PCR, and the results showed that miR-125a-5p was significantly upregulated in the agomiR-125a-5p group in comparison with agomiR-NC group (Figure 2A), which indicated that miR-125a-5p was successfully elevated in the HCT116 and HT29 cells. CCK-8 assay was used to evaluate the proliferative ability of CRC cells, and the results showed that upregulation of miR-125a-5p significantly inhibited the proliferation of HCT116 and HT29 cells compared with agomiR-NC-transfected CRC cells (Figure 2B). Furthermore, to assess the effect of miR-125a-5p on migration and invasion, wound healing and transwell assays were performed, respectively, and results suggested that miR-125a-5p overexpression suppressed cell migration (Figure 2C) and invasion (Figure 2D) in HCT116 and HT29 cells. Additionally, the effect of miR-125a-5p on CRC angiogenesis was detected using tube formation assay, as shown in Figure 2E; conditioned media (CM) from HCT116 or HT29 cells transfected with agomiR-125-5p significantly inhibited tube formation compared with CM from HCT116 or HT29 cells transfected with agomiR-NC. Together, our data indicated that miR-125a-5p suppressed CRC proliferation, migration, invasion, and angiogenesis.
Inhibition of miR-125a-5p increased the ability of CRC cell proliferation, migration, invasion, and angiogenesis
To further test the biological function of miR-125a-5p in the initiation and progression of CRC, we downregulated miR-125a-5p expression in HCT116 cells using antagomiR-125a-5p, and the efficiency of transfection was detected using qRT-PCR (Figure S1A). Our results showed that miR-125a-5p downregulation markedly reduced the ability of CRC cell proliferation, migration, and invasion and HUVEC tube formation (Figure S1B-E).
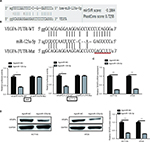
VEGFA is a direct target gene of miR-125a-5p
To explore the molecular mechanism by which miR-125a-5p exerts its functional effects on CRC cells, we predicted potential target genes by MicroRNA.org (http://www.microrna.org/) and found conserved putative miR-125a-5p binding sites at the 3′UTR of VEGFA (Figure 3A). Next, dual-luciferase reporter assay was used to verify this hypothesis, and the results showed that miR-125a-5p overexpression significantly decreased the luciferase activity of WT-VEGFA-3′UTR in HCT116 and 293T cells, whereas it failed to repress the Mut-VEGFA-3′UTR (Figure 3B). To determine whether miR-125a-5p regulates endogenous VEGFA expression in CRC cells, the mRNA and protein expression levels were detected using qRT-PCR and Western blot, respectively. miR-125a-5p overexpression was found to result in an obvious decrease in VEGFA mRNA and protein levels in HCT116 and HT29 cells (Figure 3C and D). Based on above-mentioned results, we could conclude that VEGFA was a direct target gene of miR-125a-5p in CRC cells.
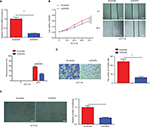
VEGFA was upregulated and inversely correlated with miR-125a-5p in CRC
VEGFA mRNA and protein levels in CRC tissues and ANTs were assessed using qRT-PCR and immunohistochemistry, respectively, and the results showed that both VEGFA mRNA and protein levels in CRC tissues were higher than those in ANTs (Figure 4A and B). Moreover, both VEGFA mRNA and protein expression levels in low miR-125a-5p CRC tissues were higher than those in high miR-125a-5p CRC tissues (Figure 4C), and Pearson’s correlation analysis further confirmed a negative correlation between miR-125a-5p expression and VEGFA mRNA or protein expression in CRC tissues (Figure 4D).
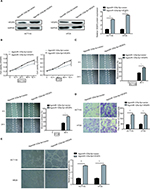
VEGFA overexpression could reverse the inhibitory effect caused by miR-125a-5p in CRC
To further explore the functional connection between miR-125a-5p and VEGFA, HCT116 and HT29 cells were cotransfected with agomiR-125a-5p and pcDNA3.1-VEGFA or blank vector. The expression of VEGFA was found to be markedly increased after cotransfection with agomiR-125a-5p and pcDNA3.1-VEGFA compared with cells transfected with agomiR-125a-5p and blank vector (Figure 5A). Overexpression of VEGFA was further shown to reverse the inhibitory effects of miR-125a-5p on cell proliferation, migration, invasion, and HUVEC tube formation in CRC (Figure 5B-E). These data revealed that miR-125a-5p inhibited cell proliferation, colony formation, migration, and HUVEC tube formation in CRC via downregulation of VEGFA.
VEGFA downregulation significantly inhibited CRC cell proliferation, migration, invasion, and HUVEC tube formation
We used siVEGFA to downregulate the expression of VEGFA in HCT116 cells, and qRT-PCR was used to detect the efficiency of transfection (Figure S2A). The results showed that VEGFA knockdown lead to decreased ability of CRC cell proliferation, migration, invasion, and HUVEC tube formation (Figure S2B-E).
miR-125a-5p inhibited tumor growth and metastasis in vivo
Given that miR-125a-5p suppressed the proliferation, migration, and invasion of CRC cells in vitro, we further explored the influence of miR-125a-5p on tumor growth and metastasis in vivo. Xenograft tumor formation in nude mice was performed to assess the in vivo role of miR-125a-5p in tumor growth, and our results showed that tumors in agomiR-125a-5p group grew much more slowly than those in agomiR-NC group (Figure 6A). Furthermore, to estimate the in vivo effects of miR-125a-5p on tumor metastasis, the tail vein of each nude mouse was injected with miR-125a-5p overexpression or negative control HCT116 cells, respectively. Histological analysis indicated that overexpression of miR-125a-5p decreased the number of lung metastatic nodules in comparison with negative group. Moreover, this difference was further verified by the examination of the lungs using H&E stains of the lung sections of mice (Figure 6B). Based on these data, we could further conclude that miR-125a-5p inhibits tumor growth and metastasis in CRC.
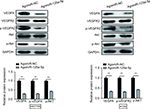
miR-125a-5p inhibited CRC progression via VEGFA/VEGFR2 signaling pathway
To further explore whether miR-125a-5p inhibited VEGFA-induced signaling pathway, we examined phosphorylation status of necessary protein kinases involved in VEGFR2 signaling pathway. As shown in Figure 7, agomiR-125a-5p significantly inhibited VEGFR2 phosphorylation and also suppressed the phosphorylation of Akt. These results revealed that miR-125a-5p might exert its inhibitory function via VEGFA/VEGFR2 signaling pathway in CRC.
Discussion
Recently, increasing number of miRNAs have been reported to be involved in CRC progression and metastasis.13-15 Bi et al reported that ectopic expression of miR-125a inhibited the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF.16 Moreover, Dai et al reported that miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A.17 These studies indicated that miRNAs might be potential diagnostic and prognostic biomarkers. Therefore, finding a novel miRNA involved in the progression of CRC becomes urgent. In our study, we found that miR-125a-5p was significantly downregulated in CRC tissues and cell lines. Moreover, decreased miR-125a-5p was markedly associated with TNM stage and poorer OS. Additionally, we verified that miR-125a-5p inhibited CRC cell proliferation, migration, invasion, and angiogenesis, whereas inhibition of miR-125a-5p promoted CRC cell proliferation, migration, invasion, and angiogenesis in vitro. In agreement with the results in vitro, we also proved that miR-125a-5p suppressed tumor growth and metastasis in vivo.
It is well known that miRNAs exert their function by regulating their target genes. As a member of VEGF family, VEGFA was associated with tumor growth, metastasis, and angiogenesis.18-20 Recently, VEGFA has been reported to be a direct target gene of many miRNAs and closely correlated with the initiation and progression in various cancers. For example, Liu et al 21showed that miR-29c acted as tumor suppressor by targeting VEGFA in lung adenocarcinoma, whereas Zhang et al22 reported that VEGFA was a direct target of miR-140-5p which suppressed progression in CRC. Ghosh et al23 demonstrated that miR-199a-3p suppressed tumor growth, metastasis, and angiogenesis via activating many oncogenes, including VEGFA. In this study, bioinformatics analysis showed that VEGFA might be a functional target gene of miR-125a-5p, and we demonstrated this hypothesis using dual-luciferase reporter assay. Furthermore, we revealed that overexpression of miR-125a-5p could reduce VEGFA both at mRNA and protein levels, and upregulation of VEGFA could reverse the decrease of proliferation, migration, invasion, and angiogenesis caused by miR-125a-5p overexpression in vitro.
Additionally, we assessed the expression of miR-125a-5p and VEGFA in CRC tissues, and an inverse correlation between the levels of VEGFA mRNA or protein expression and the levels of miR-125a-5p was noted.
VEGFA could bind to its receptor VEGFR2 and is closely associated with tumor growth, metastasis, and angiogenesis.24, 25 Here, we found that miR-125a-5p decreased the expression of VEGFA and inhibited VEGFR2 and Akt phosphorylation. Therefore, it is reasonable for us to conclude that miR-125a-5p suppressed CRC progression via VEGFA/VEGFR2 signaling pathway.
In summary, we demonstrated in our study that miR-125a-5p expression was downregulated in CRC tissues and cell lines, and that downregulated miR-125a-5p in CRC was closely associated with TNM stage and poorer OS. Moreover, we also showed upregulation of miR-125a-5p inhibited tumor growth and metastasis in vitro and in vivo, while overexpression of VEGFA could reverse the inhibitory effect caused by miR-125a-5p overexpression. Our study therefore indicated that miR-125a-5p might be a promising therapeutic target for CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9-29. | ||
Lan YT, Yang SH, Chang SC, et al. Analysis of the seventh edition of American Joint Committee on colon cancer staging. Int J Colorectal Dis. 2012;27(5):657-663. | ||
Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015;5(10):1122-1143. | ||
Liu Y, Liu R, Yang F, et al. miR-19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16(1):53. | ||
Chen X, Liu X, He B, et al. MiR-216b functions as a tumor suppressor by targeting HMGB1-mediated JAK2/STAT3 signaling way in colorectal cancer. Am J Cancer Res. 2017;7(10):2051-2069. | ||
Ye L, Jiang T, Shao H, et al. miR-1290 is a biomarker in DNA-mismatch-repair-deficient colon cancer and promotes resistance to 5-fluorouracil by directly targeting hMSH2. Mol Ther Nucleic Acids. 2017;7:453-464. | ||
Liang L, Gao C, Li Y, et al. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8(8):e2968. | ||
Yuan J, Xiao G, Peng G, et al. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457(2):171-176. | ||
Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055-1067. | ||
Hashiguchi Y, Nishida N, Mimori K, et al. Down-regulation of miR-125a-3p in human gastric cancer and its clinicopathological significance. Int J Oncol. 2012;40(5):1477-1482. | ||
Nishida N, Mimori K, Fabbri M, et al. MicroRNA-125a-5p is an independent prognostic factor in gastric cancer and inhibits the proliferation of human gastric cancer cells in combination with trastuzumab. Clin Cancer Res. 2011;17(9):2725-2733. | ||
Jiang L, Huang Q, Zhang S, et al. Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318. | ||
Vychytilova-Faltejskova P, Merhautova J, Machackova T, et al. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017;6(11):5. | ||
Huang L, Cai JL, Huang PZ, et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996-2008. | ||
Ma F, Zhang L, Ma L, et al. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exp Clin Cancer Res. 2017;36(1):158. | ||
Bi Q, Tang S, Xia, et al. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One. 2012;7(6):e40169. | ||
Dai J, Wang J, Yang L, Xiao Y, Ruan Q. miR-125a regulates angiogenesis of gastric cancer by targeting vascular endothelial growth factor A. Int J Oncol. 2015;47(5):1801-1810. | ||
Wu Y, Hooper AT, Zhong Z, et al. The vascular endothelial growth factor receptor (VEGFR-1) supports growth and survival of human breast carcinoma. Int J Cancer. 2006;119(7):1519-1529. | ||
Paez-Ribes M, Allen E, Hudock J, et al. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15(3):220-231. | ||
Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13(12):871-882. | ||
Liu L, Bi N, Wu L, et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16(1):50. | ||
Zhang W, Zou C, Pan L, et al. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37(3):1123-1133. | ||
Ghosh A, Dasgupta D, Ghosh A, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8(3):e2706. | ||
Trinh XB, Tjalma WA, Vermeulen PB, et al. The VEGF pathway and the AKT/mTOR/p70S6K1 signalling pathway in human epithelial ovarian cancer. Br J Cancer. 2009;100(6):971-978. | ||
Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114-127. |
Supplementary materials
  | Table S1 The sequences of siZFAS1, siVEGFA-NC, antagomiR-125a-5p, antagomir-NC, agomiR-125a-5p, and antagomiR-125a-5p |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.