Back to Journals » OncoTargets and Therapy » Volume 12
miR-125a-5p inhibits colorectal cancer cell epithelial–mesenchymal transition, invasion and migration by targeting TAZ
Authors Tang L, Zhou L, Wu S, Shi X, Jiang G , Niu S, Ding D
Received 17 October 2018
Accepted for publication 26 February 2019
Published 7 May 2019 Volume 2019:12 Pages 3481—3489
DOI https://doi.org/10.2147/OTT.S191247
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Gaetano Romano
Lei Tang,1 Li Zhou,2 Shengchun Wu,1 Xiaoming Shi,1 Guangwei Jiang,1 Shuai Niu,1 Dianzhu Ding1
1Vascular Surgery, Hebei General Hospital, Shijiazhuang, Hebei 050000, People’s Republic of China; 2Department of Pneumology, The Third Hospital of Hebei Medical University, Shijiazhuang, Hebei 050000, People’s Republic of China
Background: miR-125a-5p regulated biological processes in various types of cancer, including colorectal cancer (CRC). TAZ, a vital transcriptional coactivators of the Hippo pathway, was found to be overexpressed in various cancers.
Objectives: This study aims to study the effect of miR-125a-5p on the progression of CRC by regulating TAZ expression.
Methods: In this study, miR-125a-5p and TAZ expression in CRC tissue and cell lines were detected by real-time polymerase chain reaction (RT-PCR). Luciferase reporter assay was applied to detect whether TAZ was a target of miR-125a-5p. Cell migration and invasion were detected in vitro by wound-healing assay and cell invasion assay. Western blot was used to detect the expression of epithelial-mesenchymal transition (EMT)-related proteins.
Findings: The results revealed downregulation of miR-125a-5p, as well as upregulation of TAZ in CRC tissue and cell lines. TAZ was identified as a direct target of miR-125a-5p, and its expression was negatively regulated by miR-125a-5p in CRC cell lines. The functional studies revealed that overexpression of miR-125a-5p inhibited the migration, invasion and EMT of CRC cells, while upregulation of TAZ reversed the inhibitory effect caused by miR-125a-5p.
Conclusion: Our data suggest that miR-125a-5p inhibits CRC cell migration, invasion and EMT by targeting TAZ. These results suggest that miR-125a-5p serves as a potential therapeutic biomarker for CRC patients.
Keywords: miR-125a-5p, TAZ, colorectal cancer, epithelial–mesenchymal transition, migration, invasion
Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed neoplastic diseases worldwide.1 Metastasis is the major cause of death for CRC patients.2,3 Tumor metastasis begins with epithelial-mesenchymal transition (EMT). EMT has been shown to play a key role in tumor migration and invasion and is associated with tumor recurrence.4 However, the potential molecular mechanism underlying CRC remains not fully elucidated. It is therefore important to understand that the molecular mechanisms of CRC progression may provide a potential target for the effective therapeutic approaches of CRC patients.
MicroRNAs are endogenous small non-coding RNAs, consisting of 18–25 nucleotides that regulate the target genes expression by binding to the 3ʹ-untranslated regions (3ʹUTRs) of their specific mRNA targets.5 It has been reported that miRNA could promote or inhibit the progression of tumors.6 For example, miRNAs are important mediators in the progression of CRC. miR-125a-5p has been shown to act as a tumor suppressor in several tumors including hepatocellular carcinoma,7 breast cancer8 and lung cancer.9 Previous study demonstrated that miR-125a-5p inhibited colon cancer cell proliferation and induced cell apoptosis via targeting BCL2, BCL2L12 and MCL1.10
TAZ is a key transducer of the Hippo tumor-suppressor pathway and regulates many biological processes, including cell EMT.11 It has been reported that TAZ is overexpressed in multiple human cancers such as breast cancer,12 non-small cell lung cancer13 and glioblastoma.14 TAZ overexpression promoted EMT and supported pancreatic cancer progression.15 TAZ has been reported as a prognostic indicator in CRC.16 However, whether miR-125a-5p regulates CRC progression via targeting TAZ has not been elucidated.
In this study, we demonstrate that miR-125a-5p is significantly downregulated in CRC tissue and CRC cell lines. Overexpression of miR-125a-5p inhibits EMT, invasion and migration of CRC cell line by directly targeting TAZ. Our results reveal that miR-125a-5p may represent a tumor suppressor in CRC progression.
Materials and methods
Patients
A total of 30 CRC tissue samples and the correspondingly matched adjacent noncancerous tissues obtained from the patients with CRC at Hebei general hospital between 2014 and 2017 were enrolled in the study. The colorectal samples were pathologically confirmed postoperatively as colorectal adenocarcinoma. Samples were taken within 10 mins of tumor excision, immediately immersed in RNAlater™ Stabilization Solution (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −80°C until use. The patients include 14 males and 16 females and the age range is 34–74 years old. Written informed consent was obtained from all patients and that this was conducted in accordance with the Declaration of Helsinki. This study was approved by the Hebei general hospital ethics committee. Patient charts were reviewed to obtain clinical data regarding age, gender, TNM stage (according to the American Joint Committee on Cancer) and tumor differentiation and death or time of the last follow-up.
Cell lines and cell culture
The human colon cancer cell lines LoVo, SW480, SW620, HCT116, HT29 and human normal colonic epithelium cell lines (NCM460) and human embryonic kidney cell line (HEK293) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). LoVo, SW480, SW620, HCT116, HT29 and NCM460 cells were incubated in RPMI-1640 medium (Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5% CO2 incubator. HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% FBS, Gibco, USA, 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C with 5% CO2 incubator.
Transfections
The miR-125a-5p mimic, mimic control and pcDNA3-TAZ plasmid (for overexpression of TAZ) were obtained from Ribobio Co., Ltd. (Guangzhou, Guangdong, China). The human CRC cell lines LoVo and SW480 cells were seeded in 6-well plates for 48 hrs. Then, cells were transiently transfected with miR-125a-5p mimic (50 nM), mimic control (100 nM) or pcDNA3-TAZ plasmid using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
Plasmid construction and dual-luciferase reporter assay
The 3ʹ-UTR regions of human TAZ mRNA containing the predicted miR-125a-5p binding site were amplified by PCR and then cloned into the pMIR-REPORT vector (Ambion, Austin, TX, USA) to generate TAZ 3ʹUTR wild type (WT) reporter plasmids. Then, mutant (MUT) plasmids were also constructed and cloned as above. For the luciferase reporter assay, HEK293 cells were seeded in 24-well plates and incubated overnight. Then, HEK293 cells were transfected with 3ʹ-UTR WT or MUT plasmid along with miR-125a-5p mimic and mimic control at a final concentration of 50 or 100 nM using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). 24 hrs after transfection, cells were harvested. The luciferase activity was measured by dual-luciferase reporter assay system (Promega) following the manufacturer’s structure and Renilla luciferase activity was used as a control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from LoVo and SW480 cells using Trizol reagent (Invitrogen, USA) and miRNeasy extraction kit (Qiagen; Germantown, MD) according to the manufacturer’s instructions. Single-stranded cDNA was synthesized with QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA). Quantitative RT-PCR (qRT-PCR) was performed in 35 cycles using SYBR Green PCR Master mix (Takara Bio, Inc., Shiga, Japan). Each cycle consisted of 30 seconds at 94°C, 30 seconds at 60°C and 30 seconds at 72°C. TaqMan Universal PCR Master mix (all from Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used for the quantification of the miR-125a-5p. RT-qPCR was performed with the Bio-Rad CFX96 real-time PCR system (Bio-Rad, Hercules, CA, USA). GAPDH and U6 were used as the internal control. The primer sequences used were as follows: miR-125a-5p forward, 5ʹ-GGTCATTCCCTGAGACCCTTTAAC-3ʹ; reverse, 5ʹ-GTGCAGGGTCCGAGGT-3ʹ. TAZ forward, 5ʹ-ACCCACCCACGATGACCCCA-3ʹ; revere, 5ʹ-GCACCCTAACCCCAGGCCAC-3ʹ; GAPDH, forward, 5ʹ-GGAGCCAAAAGGGTCATCAT-3ʹ; revere, 5ʹ-GTGATGGCATGGACTGTGGT-3ʹ. U6, forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. The qRT-PCR experiments were performed in 3 independent experiments, each of which used 3 independent samples. 2−∆∆Ct method was used to calculate the relative expression of each gene.
Wound-healing assay
LoVo and SW480 cells (3×105 per well) were seeded in 6-well plates and transfected with miR-125a-5p mimics or co-transfected with miR-125a-5p mimic and pcDNA3-TAZ plasmid. After 48 hrs of transfection, monolayer cells were scraped using a 10 µl sterile pipette tip. Next, the migration of cells was observed at 0 hrs and 24 hrs after wounding using a phase contrast microscope (IX711; Olympus, Tokyo, Japan). Quantitative analysis of the wound-healing area was done using Image J software (National Institutes of Health, MA, USA).
Cell invasion assay
Cell invasion assay was performed in 24-well invasion chambers containing a transwell membrane filter (Corning, NY, USA). After 48 hrs transfection, LoVo and SW480 cells (3×104 cells/well) were added in the upper chamber of the Transwells, in which the porous membrane was either coated with Matrigel (BD Bioscience). 500 µL of 10% FBS medium was placed in the lower chamber. Incubation was carried out for 24 hrs at 37°C; no migrated cells on the upper membrane were removed using cotton swabs. The number of invading cells was used to assess invasive ability. Cells' invasion number was quantitated from five random fields for each chamber under an Olympus microscope (Olympus).
Western blot analysis
Total protein from transfected LoVo and SW480 cells was extracted using protein lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Bicinchoninic acid assay kit (Beyotime Institute of Biotechnology) was used to measure the protein concentration (Beyotime Institute of Biotechnology). Tissue lysate (30 µg) was subjected to Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels (Sigma-Aldrich) and transferred to polyvinylidene difluoride membranes (PVDF) (EMD Millipore, Billerica, MA, USA). Blots were blocked with 5% (w/v) skimmed milk in TBST for 1 hr at room temperature and then incubated with primary antibodies anti-TAZ (cat no. ab110239, 1:1,000, Abcam, Cambridge, UK), anti-E-cadherin (cat no. 14472, 1:1,000, Cell Signaling Technology, Danvers, MA, USA), anti-N-cadherin (cat no. 14215, 1:1,000, Cell Signaling Technology), anti-β-catenin (cat no. 8480, 1:1,000, Cell Signaling Technology) and anti-vimentin (cat no. 5741, 1:1,000, Cell Signaling Technology), anti-GAPDH (14C10) (cat no. #2118, 1:1,000, Cell Signaling Technology) overnight at 4°C. Next, the membranes were incubated with the secondary antibody HRP-linked anti-rabbit IgG antibody (cat no. #7074, 1:2,000, Cell Signaling Technology) for 1 hr at room temperature. Proteins bands were detected using enhanced chemiluminescence detection system (GE Healthcare Life Sciences, Chalfont, UK) following the manufacturer’s instructions. GAPDH was used as a positive control.
Statistical analysis
Statistical analysis was performed using SPSS 15.0 software (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance (ANOVA) was used to assess the statistical differences between the groups. Correlations between clinicopathological characteristics and miR-125a-5p expression in patients with retinoblastoma were analyzed using the Pearson’s Chi-square test. The data were determined as the mean ± standard deviation. P<0.05 was considered statistically significant.
Results
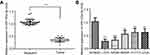
miR-125a-5p expression is down-regulated in CRC tissues and cell lines
In order to investigate the role of miR-125a-5p in CRC development, the levels of miR-125a-5p in CRC tissues and CRC cell lines were detected by qRT-PCR. qRT-PCR results showed that the miR-125a-5p expression was significantly downregulated in CRC tissues compared with adjacent tissues (Figure 1A). Moreover, we selected five commonly used CRC cell lines (LoVo, SW620, SW480, HCT116 and HT29) to detect the levels of miR-125a-5p in CRC cell lines. The results showed that the mRNA levels of miR-125a-5p were significantly downregulated in all five CRC cell lines compared with normal colonic mucosa epithelial cells (NCM460) (Figure 1B). These results indicate that miR-125a-5p could involve in the development of CRC. In addition, we evaluated the correlation between expression levels of miR-125a-5p with clinical parameters (Table 1) and found that miR-125a-5p downregulation was significantly associated with TNM stage and invasion and migration.
 | Table 1 Correlation between clinicopathological characteristics and miR-125a-5p expression in patients with colorectal cancer |
TAZ is a direct target gene of miR-125a-5p
In order to identify the potential targets of miR-125a-5p, the prediction programs PicTar (
miR-125a-5p inhibits migration and invasion of CRC cells via directly targeting TAZ
To evaluate the effects of miR-125a-5p on CRC and whether this effect is mediated by its targets TAZ gene, LoVo and SW480 cells were transfected with miR-125a-5p mimic, mimic control or co-transfected with miR-125a-5p mimic and TAZ overexpression plasmids. Our results showed that TAZ overexpression plasmids significantly increased the expression of TAZ (Figure 3A and B). In addition, wound healing (Figure 3C) and cell invasion assays (Figure 3D) showed that miR-125a-5p mimic transfection significantly suppressed the migration and invasion of LoVo and SW480 cells; restoration of TAZ was able to partly overcome the inhibitory effects of miR-125a-5p on cell migration and invasion. Together, these results indicated that miR-125a-5p inhibits migration and invasion of CRC cells by directly targeting TAZ.
miR-125a-5p inhibits epithelial–mesenchymal transition of CRC cells via directly targeting TAZ
To detect the effect of miR-125a-5p on EMT of CRC cells and whether this effect is mediated by its targets TAZ gene, the expression of several well-characterized EMT molecules was performed by Western blot. As shown in Figure 4A and B, transfection of miR-125a-5p significantly increased the levels of E-cadherin while decreased the levels of N-cadherin, vimentin and β-catenin in LoVo and SW480 cells. Over-expression of TAZ could reverse the effects of miR-125a-5p on EMT of CRC cells. Together, these results indicated that miR-125a-5p inhibits CRC cells EMT by directly targeting TAZ.
Discussion
CRC is one of the most common cancers, posing a serious demographic and economic burden worldwide.1 Recent study has shown that miRNA participates in CRC process.17 The findings of our study demonstrated that miR-125a-5p expression was down-regulated in CRC tissues and cell lines. We also determined that miR-125a-5p inhibited cell epithelial–mesenchymal transition, invasion and migration in CRC by directly suppressing TAZ expression.
Previous studies have reported that miR-125a-5p functions as a tumor suppressor in a variety of cancers, including CRC10,18 For example, miR-125a-5p functions as a tumor suppressor in breast cancer by downregulating BAP1.19 miR-125a-5p has been reported to inhibit invasion and metastasis of gastric cancer cells by targeting BRMS1 expression.18 In addition, miR-125a-5p suppressed CRC progression by targeting VEGFA.20 miR-125a-5p inhibited colon cancer cell proliferation and induced apoptosis via targeting BCL2, BCL2L12 and MCL1.10 In the present study, we found that miR-125a-5p was downregulated in CRC tissue and cells. Overexpression of miR-125a-5p inhibited EMT, invasion and migration of CRC cells.
TAZ is a key functional component of the Hippo pathway and plays vital roles in tumor proliferation and invasion.21 TAZ expression has been reported as a prognostic indicator in CRC.16 TAZ has also been reported to regulate mesenchymal stem cell differentiation.22 TAZ promotes epithelial to mesenchymal transition in neuroblastoma cells.23 In this study, we found TAZ was a direct target of miR-125a-5p, which was consistent with previous study.11 Moreover, miR-125a-5p and TAZ expression were inversely correlated in CRC tissue and cell lines. TAZ expression was downregulated in CRC tissue and cell lines in which miR-125a-5p was overexpressed. Furthermore, CRC cell EMT, invasion and migration capacity in the miR-125a-5p mimic group were restored by TAZ overexpression.
In summary, our study demonstrate that miR-125a-5p represses CRC cell EMT, invasion and migration by targeting TAZ. These evidence suggests that TAZ and miR-125a-5p may serve as promising diagnostic and therapeutic targets for patients with CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi:10.1002/ijc.29210
2. Van Cutsem E, Cervantes A, Nordlinger B, Arnold D. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):i1–i9. doi:10.1093/annonc/mdu061.1
3. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi:10.3322/caac.21220
4. Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol Res Pract. 2015;211:557–569. doi:10.1016/j.prp.2015.05.010
5. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi:10.1016/j.ydbio.2006.08.028
6. Bhartiya D, Scaria V. Genomic variations in non-coding RNAs: structure, function and regulation. Genomics. 2016;107:59–68. doi:10.1016/j.ygeno.2016.01.005
7. Kim JK, Noh JH, Jung KH, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi:10.1002/hep.26101
8. Sun Y, Liu X, Zhang Q, et al. Oncogenic potential of TSTA3 in breast cancer and its regulation by the tumor suppressors miR-125a-5p and miR-125b. Tumour Biol. 2016;37:4963–4972. doi:10.1007/s13277-015-4178-4
9. Wang G, Mao W, Zheng S, Ye J. Epidermal growth factor receptor-regulated miR-125a-5p–a metastatic inhibitor of lung cancer. FEBS J. 2009;276:5571–5578. doi:10.1111/j.1742-4658.2009.07238.x
10. Tong Z, Liu N, Lin L, Guo X, Yang D, Zhang Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi:10.1016/j.biopha.2015.07.036
11. Yuan J, Xiao G, Peng G, et al. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457:171–176. doi:10.1016/j.bbrc.2014.12.078
12. Chan SW, Lim CJ, Guo K, et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 2008;68:2592–2598. doi:10.1158/0008-5472.CAN-07-2696
13. Malik SA, Khan MS, Dar M, Hussain MU, Mudassar S. TAZ is an independent prognostic factor in non-small cell lung carcinoma: elucidation at protein level. Cancer Biomark. 2017;18:389–395. doi:10.3233/CBM-160263
14. Yang R, Wu Y, Zou J, et al. The Hippo transducer TAZ promotes cell proliferation and tumor formation of glioblastoma cells through EGFR pathway. Oncotarget. 2016;7:36255–36265. doi:10.18632/oncotarget.9199
15. Xie D, Cui J, Xia T, et al. Hippo transducer TAZ promotes epithelial mesenchymal transition and supports pancreatic cancer progression. Oncotarget. 2015;6:35949–35963. doi:10.18632/oncotarget.v6i34
16. Yuen H-F, McCrudden CM, Huang Y-H, et al. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One. 2013;8:e54211. doi:10.1371/journal.pone.0054211
17. Xu P, Wang J, Sun B, Xiao Z. Integrated analysis of miRNA and mRNA expression data identifies multiple miRNAs regulatory networks for the tumorigenesis of colorectal cancer. Gene. 2018;659:44–51. doi:10.1016/j.gene.2018.03.050
18. Cao Y, Tan S, Tu Y, et al. MicroRNA-125a-5p inhibits invasion and invasion and migration of gastric cancer cells by targeting BRMS1 expression. Oncol Lett. 2018;15:5119–5130. doi:10.3892/ol.2018.7983
19. Yan L, Yu M-C, Gao G-L, et al. MiR-125a-5p functions as a tumour suppressor in breast cancer by downregulating BAP1. J Cell Biochem. 2018;119(11):8773–8783. doi:10.1002/jcb.27124
20. Yang X, Qiu J, Kang H, Wang Y, Qian J. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res. 2018;10:5839–5853. doi:10.2147/CMAR.S161990
21. Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi:10.1016/j.devcel.2010.09.011
22. Hong J-H, Hwang ES, McManus MT, et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi:10.1126/science.1110955
23. Wang Q, Xu Z, An Q, et al. TAZ promotes epithelial to mesenchymal transition via the upregulation of connective tissue growth factor expression in neuroblastoma cells. Mol Med Rep. 2015;11:982–988. doi:10.3892/mmr.2014.2818
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.