Back to Journals » OncoTargets and Therapy » Volume 13
MiR-1193 Inhibits the Malignancy of Cervical Cancer Cells by Targeting Claudin 7 (CLDN7)
Authors Zhang B , Lin Y, Bao Q, Zheng Y, Lan L
Received 23 January 2020
Accepted for publication 2 May 2020
Published 19 May 2020 Volume 2020:13 Pages 4349—4358
DOI https://doi.org/10.2147/OTT.S247115
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Bin Zhang,* Yao Lin,* Qiufang Bao, Yantong Zheng, Lan Lan
Department of Obstetrics and Gynecology, The First Affiliated Hospital of Fujian Medical University, Fujian 350005, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bin Zhang Email [email protected]
Objective: MicroRNAs (miRNAs) are highly involved in cancer development, including in cervical cancer (CC). In this study, we aimed to investigate the role and possible mechanism of a poorly studied miRNA, miR-1193, in CC progression.
Materials and Methods: Expression of miR-1193 was determined in 60 pairs of cervical samples. The impacts of miR-1193 on CC cell proliferation, invasion and migration capacities were verified by CCK-8, transwell and wound healing assays, respectively. Then, bioinformatics prediction, luciferase reporter assay, qRT-PCR and Western blot were successively conducted to study the targeting of claudin 7 (CLDN7) by miR-1193. After CLDN7 was restored in miR-1193-overexpressed cells, the rescue effects were determined. Finally, CLDN7 expression was analyzed in cervical samples, and its expression correlation with miR-1193 was explored.
Results: Compared with paired normal tissues, miR-1193 was sharply decreased in abnormal tissues (intraepithelial lesions and cancerous tissues). Especially, miR-1193 expression was gradually decreased in low-grade squamous intraepithelial lesions, high-grade squamous intraepithelial lesions and CC. Enforced expression of miR-1193 inhibited CC cell proliferation, invasion and migration. Mechanistically, we confirmed CLDN7 as a target of miR-1193, and restoration of CLDN7 robustly rescued the tumor suppressing effects of miR-1193 in CC cells. CLDN7 was upregulated in abnormal cervical tissues and its expression exhibited inverse correlation with that of miR-1193 in CC.
Conclusion: Our results suggested that miR-1193 exerted tumor inhibitory roles in CC malignancy by directly targeting CLDN7.
Keywords: cervical cancer, miR-1193, tumor suppressing, CLDN7
Introduction
Cervical cancer (CC) ranks as one of the most common and lethal malignancies in females worldwide, which greatly imperils the physical and mental health of women, and the situation is worse in developing countries.1,2 Persistent infection of the human papillomavirus (HPV) is a key attribute but not adequate to bring about cervical carcinogenesis.3 Although extensive investigation has broaden our knowledge on some mechanism of CC pathogenesis, unfortunately, the current 5-year survival rate of CC patients at the advanced stages remains dissatisfactory.4,5 Therefore, it is urgent to identify more efficient therapeutic targets and develop novel strategies for treatment of this disease.
MicroRNAs (miRNAs) are a large family of short (~22 nucleotides in length) non-coding RNAs in eukaryotes. Through directly binding to the complementary sequences in the 3ʹ-untranslated regions (3ʹ-UTRs) of their target mRNAs and leading to the target mRNA degradation and/or translation suppression, miRNAs are deemed as crucial regulators of gene expression and highly implicated in tumorigenesis, including in CC development.6–8 Numerous miRNAs have been identified as oncogenes or tumor suppressors in CC, such as miR-338-3p,9 miR-21110 and miR-877.11 Besides, some miRNAs have been reported to be abnormally expressed and have potential diagnostic or prognostic significance in clinic.12 Recently, miR-1193, a newly found and poorly studied miRNA, was found to be dysregulated in cutaneous squamous cell carcinoma, T-cell leukemia and breast cancer.13–15 However, its implication in CC remains unknown.
In this study, we examined the expression, function and mechanism of miR-1193 in CC progression. Our results indicated that miR-1193 was gradually downregulated in LSIL (low-grade squamous intraepithelial lesions), HSIL (high-grade squamous intraepithelial lesions) and CC tissues. Gain-of-function and mechanistic studies revealed that miR-1193 acted as a tumor suppressor by targeting claudin 7 (CLDN7) in CC cells.
Materials and Methods
Patients and Tissues
Sixty pairs of human cervical samples including abnormal cervical tissues (LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions and CC) and matched normal tissues (more than 5cm from abnormal tissues were necessary) were collected from Jan 2018 to Dec 2019 in First Affiliated Hospital of Fujian Medical University 350005 PR China. They were all freshly frozen in liquid nitrogen until used. All patients had not undergone preoperative chemotherapy or radiotherapy treatment. All patients had signed informed consent and Ethics Committee approval was given by First Affiliated Hospital of Fujian Medical University 350005 PR China.
Cell Culture
Six cervical cancer cell lines (C-33A, CaSki, HCE1, Hela, Hela 229 and SiHa) and one normal human cervix epithelial cell line (Ect1/E6E7) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Life Technology, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Life Technology, Thermo Fisher Scientific, Waltham, MA, USA), 100U/mL penicillin and 100μg/mL streptomycin (P/S, Life Technology, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C with 5% CO2 in a humidified incubator.
MiRNA Mimics and Transfections
MicroRNA negative controls (miR-C) and miR-1193 mimics (miR-1193) were purchased from GenePharma Company (Shanghai, P. R. China). Transfections were performed using Lipofectamine™ 2000 Transfection Reagent (Life Technology, Thermo Fisher Scientific, Waltham, MA, USA) in accordance with the manufacturer’s instructions.
qRT-PCR (Quantitative Real-Time Polymerase Chain Reaction)
Total RNA was extracted from paired cervical cancer tissues and cultured cells with TRIzol® reagent (Life Technology, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The relative level of miR-1193 and U6B was detected using a SYBR® PrimeScript® miRNA RT-PCR Kit (TaKaRa, Dalian, Liaoning, P. R. China) according to the manufacturer’s instructions. For detection of CLDN7 and GAPDH mRNA expression, the first-strand cDNA was synthesized using M-MLV Reverse Transcriptase (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. qRT-PCR was conducted using SYBR Premix Ex Taq (TaKaRa, Dalian, Liaoning, China) in the ABI 7900 Fast system (Applied Biosystems, Foster City, CA, USA). The relative expression of miR-1193 and CLDN7 was normalized to U6B and GAPDH, respectively, by using the 2−ΔΔCT method.
CCK-8 Assay
24h after transfection, Hela or SiHa cells were resuspended and seeded into 96-well plates at a density of 2.5×103 cells/well. Cell proliferation ability was evaluated by conducting the CCK-8 assay (Dojindo, Kumamoto, Japan) at four time points: Day1, Day2, Day3 and Day4 after incubation. A total of 10μL CCK-8 reagent was added to each well and incubated at 37°C with 5% CO2 for another 2h. Finally, the optical density of each well was measured at a wavelength of 450nm using a microplate reader (Molecular Device, San Jose, CA, USA).
Transwell Assay
Cell invasion ability was determined using transwell assay. Briefly, 2.5×105 cells were seeded into the upper chamber of 24-well Transwell chambers (Corning Incorporated, Corning, NY, USA) with a pore size of 8.0μm. In the lower chamber, 600μL DMEM medium with 10% FBS was added. 24h later, the non-invading cells on the upper surface of the filter were gently removed with a cotton swab, whereas the cells that invaded into the lower surface of the filter were fixed with paraformaldehyde for 30min and stained with 0.1% crystal violet for 20min at room temperature. The cells were photographed with an inverted light microscope (Olympus, Tokyo, Japan) for at least five individual fields.
Wound Healing Assay
Cell migration ability was determined using wound-healing assay. Controls (miR-C) or miR-1193 mimics (miR-1193) transfected cells (2.5×105/well) were cultured in 6-well plates for 24h. Subsequently, the cells were scratched with a sterile plastic micropipette tip to generate a wound in the monolayer. The cells were washed twice with PBS and cultured in DMEM medium without serum for 24h. Then, the wound was imaged at 0h and 24h under an inverted microscope (Olympus, Tokyo, Japan), and the separation distance between wound sides was measured from the images by ImageJ software at three different sites of the wound area of gaps. Three independent experiments were performed and the wound area values were averaged. The result was shown as the ratio of experimental group to control group.
Plasmid Construction
The wild type fragment of CLDN7 3ʹ-UTR-WT containing the miR-1193 predicted binding site or mutant fragment of CLDN7 3ʹ-UTR-MT was synthesized by Sangon Biotech Co., Ltd. (Shanghai, P. R. China). The two fragments were sub-cloned into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega Corporation, Madison, WI, USA). The coding sequence of CLDN7 mRNA was amplified by PCR using KOD-plus-Ver.2 kit (TOYOBO, Osaka, Japan) and subcloned into pcDNA™3.1/myc-His A Mammalian Expression Vectors (Life Technology, Thermo Fisher Scientific, Waltham, MA, USA).
Luciferase Reporter Assay
Hela or SiHa cells were co-transfected with controls (miR-C) or miR-1193 mimics (miR-1193) and luciferase reporter vectors of the wild type CLDN7 3ʹ-UTR (CLDN7 3ʹ-UTR-WT) or mutant CLDN7 3ʹ-UTR (CLDN7 3ʹ-UTR-MT). 48h later, luciferase activity was detected using Dual-Luciferase® Reporter Assay System (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions.
Western Blot
Briefly, cells were lysed using radioimmunoprecipitation lysis buffer (RIPA buffer, Beyotime, Nanjing, Jiangsu, China). The protein concentration was determined using a BCA protein assay kit (Beyotime, Nanjing, Jiangsu, China). An equal amount (25μg) protein samples were separated on 10% SDS-PAGE, then the proteins were transferred to nitrocellulose (NC) membrane (Millipore, Billerica, MA). The membranes were blocked by 5% skim milk for 1h at room temperature. Then, the membranes were incubated with antibodies against CLDN7, IGF2BP2, β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA) or TM9SF3 (Abclonal Technology, Wuhan, China) overnight at 4°C, followed by HRP-linked secondary antibodies (Beyotime, Nanjing, Jiangsu, China) 1h at room temperature. Finally, the protein blots were detected by the enhanced chemiluminescence detection system (Pierce, Billerica, MA). β-actin was used as an internal control.
Target Prediction
Targets of miR-1193 were searched on Targetscan Release 7.2 (http://www.targetscan.org/vert_72/), and the results suggested CLDN7 as a potential target of miR-1193. To further confirm whether CLDN7 was directly targeted by miR-1193, we obtained more information about the sequence of CLDN7 3ʹ-UTR on Targetscan.
Statistical Analysis
Statistical analyses were performed by GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, USA). Statistical significance of difference between two groups was determined by a two-tailed Student’s t-test. Statistical significance of difference between three or more groups was determined by one-way ANOVA. The correlation between miR-1193 and CLDN7 expressions was analyzed by Spearman correlation. All experiments were performed in triplicate and presented as mean ± SD. P value of <0.05 was considered as statistically significant.
Results
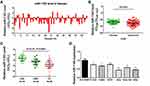
Decreased Expression of MiR-1193 in CC Tissues and Cell Lines
To characterize the role of miR-1193 in CC, we firstly explored its expression pattern in clinical cervical samples. In the 60 pairs of cervical samples collected (including LSIL, HSIL, CC and matched normal tissues), qRT-PCR result showed that miR-1193 was frequently downregulated in abnormal tissues (LSIL, HSIL and CC) compared with matched normal tissues (Figure 1A and B). Furthermore, when classifying these clinical cervical samples with LSIL, HSIL and CC, the statistical analysis result showed that miR-1193 was more decreased in CC (Figure 1C), indicating the relevance of miR-1193 in CC progression. Meanwhile, we also detected the expression of miR-1193 in a panel of CC cell lines. The result showed that in comparison with the normal cervix epithelial cell line (Ect1/E6E7), miR-1193 was downregulated in the six cervical cancer cell lines (C-33A, CaSki, HCE1, Hela, Hela 229, SiHa, Figure 1D). Therefore, these expression detection results confirmed that miR-1193 was decreased in CC and might be highly involved in CC progression.
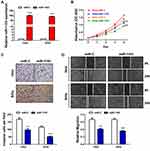
The Inhibitory Roles of MiR-1193 in CC
Then, we overexpressed miR-1193 in two CC cell lines (Hela and SiHa), which contained relatively lower endogenous level of miR-1193 (Figure 1D). After validation of the overexpression in the two cell lines by qRT-PCR (Figure 2A), we performed CCK-8 assay to evaluate the impact of miR-1193 in cell proliferation. The results shown in Figure 2B demonstrated that miR-1193 significantly inhibited CC cell proliferation in these two cell lines. In addition, its enforced expression markedly inhibited CC cell invasion and migration (Figure 2C and D). These tumor suppressing activities of miR-1193 are very similar to those in breast cancer and T-cell leukemia.14,15
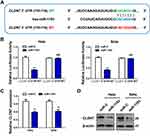
MiR-1193 Directly Targets CLDN7 in CC Cells
Through the above expression and function studies, we concluded that miR-1193 was an important tumor suppressing miRNA in CC. However, the mechanism about how miR-1193 exerted tumor suppressing activities was unknown in CC. As we known, miRNAs negatively regulated the target gene expression in a post-transcription manner. We utilized Targetscan to predict the possible targets of miR-1193. As shown in Figure 3A, we found an interaction between miR-1193 and 3ʹ-UTR of claudin 7 (CLDN7). Luciferase reporter assay revealed that miR-1193 overexpression led to the reduction of the wild type reporter (CLDN7 3ʹ-UTR-WT) activities in the two CC cell lines, whereas miR-1193 overexpression had nearly no impact on the mutant type reporter (CLDN7 3ʹ-UTR-MT, Figure 3A and B). Furthermore, qRT-PCR and Western blot experiments showed that miR-1193 significantly repressed the mRNA and protein expression of CLDN7 (Figure 3C and D), but IGF2BP2 and TM9SF3, another two targets of miR-1193 reported in breast cancer and T-cell leukemia, respectively, showed minor or almost no change at both mRNA and protein levels (Supplemental Figure 1). Therefore, we confirmed that CLDN7 was a direct target of miR-1193 in CC cells.
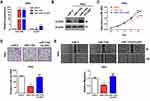
Restoration Effects of CLDN7 in miR-1193-Overexpressed CC Cells
To investigate whether miR-1193 exerted tumor suppressing effects via targeting CLDN7 in CC cells, we co-transfected CLDN7 expressing plasmid with miR-1193 mimics, and compared the proliferation, invasion and migration capacities with the single miR-1193 mimics transfection in Hela cells. qRT-PCR and Western blot both validated the successful transfection of CLDN7 (Figure 4A and B). We found that restoration of CLDN7 robustly rescued the tumor suppressing effects of miR-1193 in CC cells, including in cell proliferation, invasion and migration (Figure 4C–E). These results clearly demonstrated that miR-1193 inhibits CC progression at least partially via downregulating CLDN7.
Increased Expression of CLDN7 in CC
Through the above studies, we learned that CLDN7 was a vital target of miR-1193 and mediated the tumor suppressing effects of miR-1193 in CC cells. However, whether CLDN7 itself was aberrantly expressed in CC remains unclear. Using the above-mentioned 60 pairs of cervical samples, we conducted qRT-PCR assay to detect its expression. As shown in Figure 5A and B, we found that CLDN7 was statistically significantly upregulated in most of abnormal tissues compared with matched normal tissues. The increase of CLDN7 was also confirmed by the mRNA expression data from the GEPIA database (http://gepia.cancer-pku.cn/; Supplemental Figure 2A) and the immunohistochemistry data from the human protein atlas (https://www.proteinatlas.org/, Figure 5C). In contrast, the upregulation of CLDN7 was more obvious in CC than HSIL, and also more obvious in HSIL than LSIL (Figure 5D). Additionally, we found that upregulated CLDN7 expression was inversely correlated with downregulated miR-1193 in these 60 pairs of cervical samples (Figure 5E). This negative correlation was further confirmed by using the online LinkedOmics (http://www.linkedomics.org/) data when analyzing the expression of miR-1193 and CLDN7 in CESC (cervical squamous cell carcinoma; Supplemental Figure 2B). Collectively, our data demonstrated that CLDN7 was a critical and functional target of miR-1193, which exhibited upregulation and might be highly implicated in CC progression.
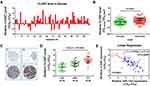 |
Figure 5 Increased expression of CLDN7 in cervical cancer. (A) Difference in the expression of CLDN7 in 60 pairs of cervical samples by qRT-PCR analysis. (B) Difference in the expression of CLDN7 in 60 pairs of abnormal cervical tissues (Abnormal) and matched normal tissues (Normal) by qRT-PCR analysis. (C) Immunohistochemistry analyses from the online tool human protein atlas (https://www.proteinatlas.org/) validated CDLN7 protein upregulation in non-cancerous cervical tissues (a, b) and cervical cancer tissues (c, d). Magnification: 100×. (D) Difference in the expression of CLDN7 in LSIL (N=16), HSIL (N=16) and CC (N=28) tissues by qRT-PCR analysis. (E) Spearman correlation analysis was performed to explore the negative correlation between miR-1193 and CLDN7 mRNA levels in 60 paired cervical samples (N=60, P<0.0001). Abbreviations: LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions; CC, cervical cancer. |
Discussion
Abnormal expressions of miRNAs are frequently reported in cancers and they are closely related to the initiation and progression of multiple human cancers.16 Importantly, various reports have established that miRNAs might be promising biomarkers or therapeutic targets for some cancers, including CC.17,18 Therefore, studies centering on the roles and mechanisms of miRNAs in CC may largely benefit the improvement of diagnosis, therapy and prognosis of this dismal disease.
MiR-1193 was firstly identified in the melanoma miRNAome by deep sequencing by Stark and his colleagues.19 To the best of our knowledge, there are rare studies investigating its roles in human cancers. Until recently, two groups reported the tumor suppressing functions of miR-1193 in breast cancer and T-cell leukemia.14,15 Their findings indicated that miR-1193 could inhibit cell proliferation and invasion in the two types of malignancies by targeting individual targets, IGF2BP2 and TM9SF3, suggesting the common inhibitory roles in tumors while specific targets of miR-1193.14,15
In our study, we examined the expression, function and possible mechanism of miR-1193 in cervical samples and cells. Since it remained unclear about the roles of miR-1193 in CC, our study presented the details for the first time. We showed that miR-1193 was especially downregulated in CC tissues. These results highly indicated that miR-1193 might take part in CC progression. Consistently, gain-of-function investigation revealed that miR-1193 inhibited CC cell proliferation, invasion and migration. The tumor inhibitory activities of miR-1193 quite conform to its downregulation in cervical samples, and these results are very similar to those in breast cancer and T-cell leukemia,14,15 suggesting the common tumor suppressing roles of miR-1193 in these malignancies.
As we known, miRNAs could regulate gene expression in a post-transcription manner. It is well recognized that one single mRNA contains several binding sites for distinct miRNAs, likewise, one single miRNA might target several target mRNAs.20 In breast cancer, miR-1193 was found to target the insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) mRNA, thus repress the downstream oncogenic ERK and PI3K/AKT pathways.15 In T-cell leukemia, transmembrane 9 superfamily 3 (TM9SF3) was validated as a target of miR-1193 and it displayed a strong negative correlation with miR-1193 in T-cell leukemia.14
In our study, we showed that miR-1193 targeted the claudin 7 (CLDN7) mRNA at the 3ʹ-UTR region, therefore, downregulated CLDN7 expression, but hardly regulate the expression of IGF2BP2 or TM9SF3, indicating that miR-1193 might have specific or at least favorite targets in different malignancies. Importantly, rescue experiments confirmed that CLDN7 could mediate the tumor suppressing effects of miR-1193 in CC cells, since CLDN7 restoration largely rescued the tumor inhibitory activities of miR-1193.
The claudin family consists over 24 members, and each of them displays a tissue-specific distribution.21 They are tetraspan transmembrane protein components of tight junctions and determine the properties of cell-cell contacts between two neighboring cells.22 To our knowledge, CLDN7 exerts both tumor promoting and tumor suppressing roles in human malignancies, depending on the tumor types. For instance, CLDN7 is downregulated in esophageal and breast cancer and functions as a tumor suppresser.23,24 However, in ovarian cancer, it is upregulated and functions as an oncogene.25 In the present study, we provide crucial evidence that CLDN7 was more upregulated in CC tissues. Although direct evidences by loss- or gain-of-function studies to characterize the function of CLDN7 in CC were absent in our study, the rescue experiments that CLDN7 restoration could recover the reduced proliferation, migration and invasion caused by miR-1193, also supported that CLDN7 itself was a vital oncogene in CC cells. Furthermore, we showed that CLDN7 expression was inversely correlated with miR-1193. This result once more validated that the targeting of CLDN7 by miR-1193.
In summary, the findings present in this study identified miR-1193 as a crucial tumor suppressor in CC by inhibiting cell proliferation, invasion and migration. We verified that CLDN7 was a direct and important target of miR-1193 in CC cells, since its restoration significantly recovered the tumor suppressing effects caused by miR-1193. Our study highlights the possible application of miR-1193 as a novel target in CC therapy.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Su SY, Huang JY, Ho CC, Liaw YP. Evidence for cervical cancer mortality with screening program in Taiwan, 1981–2010: age-period-cohort model. BMC Public Health. 2013;13:13. doi:10.1186/1471-2458-13-13
3. Banister CE, Liu C, Pirisi L, Creek KE, Buckhaults PJ. Identification and characterization of HPV-independent cervical cancers. Oncotarget. 2017;8(8):13375–13386. doi:10.18632/oncotarget.14533
4. Castelnau-Marchand P, Chargari C, Maroun P, et al. Clinical outcomes of definitive chemoradiation followed by intracavitary pulsed-dose rate image-guided adaptive brachytherapy in locally advanced cervical cancer. Gynecol Oncol. 2015;139(2):288–294. doi:10.1016/j.ygyno.2015.09.008
5. Bosch FX, Lorincz A, Muñoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. J Clin Pathol. 2002;55(4):244–265. doi:10.1136/jcp.55.4.244
6. Nahand JS, Taghizadeh-Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–17099. doi:10.1002/jcp.28457
7. Wang JY, Chen LJ. The role of miRNAs in the invasion and metastasis of cervical cancer. Biosci Rep. 2019;39(3):
8. Srivastava SK, Ahmad A, Zubair H, et al. MicroRNAs in gynecological cancers: small molecules with big implications. Cancer Lett. 2017;407:123–138. doi:10.1016/j.canlet.2017.05.011
9. Hua FF, Liu SS, Zhu LH, et al. MiRNA-338-3p regulates cervical cancer cells proliferation by targeting MACC1 through MAPK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(23):5342–5352. doi:10.26355/eurrev_201712_13919
10. Qu X, Gao D, Ren Q, Jiang X, Bai J, Sheng L. miR-211 inhibits proliferation, invasion and migration of cervical cancer via targeting SPARC. Oncol Lett. 2018;16(1):853–860. doi:10.3892/ol.2018.8735
11. Meng F, Ou J, Liu J, et al. MicroRNA-877 is downregulated in cervical cancer and directly targets MACC1 to inhibit cell proliferation and invasion. Exp Ther Med. 2019;18(5):3650–3658. doi:10.3892/etm.2019.7989
12. Pardini B, De Maria D, Francavilla A, Di Gaetano C, Ronco G, Naccarati A. MicroRNAs as markers of progression in cervical cancer: a systematic review. BMC Cancer. 2018;18(1):696. doi:10.1186/s12885-018-4590-4
13. Wang J, Li C, Xu L, Yang C, Zhang X. MiR-1193 was sponged by LINC00963 and inhibited cutaneous squamous cell carcinoma progression by targeting SOX4. Pathol Res Pract. 2019;215(10):152600. doi:10.1016/j.prp.2019.152600
14. Shen L, Du X, Ma H, Mei S. miR-1193 suppresses the proliferation and invasion of human T-cell leukemia cells through directly targeting the transmembrane 9 superfamily 3 (TM9SF3). Oncol Res. 2017;25(9):1643–1651. doi:10.3727/096504017X14908284471361
15. Li X, Li Y, Lu H. miR-1193 suppresses proliferation and invasion of human breast cancer cells through directly targeting IGF2BP2. Oncol Res. 2017;25(4):579–585. doi:10.3727/97818823455816X14760504645779
16. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
17. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi:10.1038/nrd.2016.246
18. Satapathy S, Batra J, Jeet V, Thompson EW, Punyadeera C. MicroRNAs in HPV associated cancers: small players with big consequences. Expert Rev Mol Diagn. 2017;17(7):711–722. doi:10.1080/14737159.2017.1339603
19. Stark MS, Tyagi S, Nancarrow DJ, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One. 2010;5(3):e9685. doi:10.1371/journal.pone.0009685
20. Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11(12):1753–1761. doi:10.1261/rna.2248605
21. Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10(8):235. doi:10.1186/gb-2009-10-8-235
22. Krause G, Winkler L, Mueller SL, Haseloff RF, Piontek J, Blasig IE. Structure and function of claudins. Biochim Biophys Acta. 2008;1778(3):631–645. doi:10.1016/j.bbamem.2007.10.018
23. Usami Y, Chiba H, Nakayama F, et al. Reduced expression of claudin-7 correlates with invasion and metastasis in squamous cell carcinoma of the esophagus. Hum Pathol. 2006;37(5):569–577. doi:10.1016/j.humpath.2005.12.018
24. Sauer T, Pedersen MK, Ebeltoft K, Naess O. Reduced expression of Claudin-7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16(4):193–198. doi:10.1111/j.1365-2303.2005.00257.x
25. Dahiya N, Becker KG, Wood WH
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.