Back to Journals » OncoTargets and Therapy » Volume 12
miR-106b promotes proliferation and invasion by targeting Capicua through MAPK signaling in renal carcinoma cancer
Authors Miao LJ, Yan S, Zhuang QF, Mao QY, Xue D, He XZ, Chen JP
Received 21 August 2018
Accepted for publication 1 February 2019
Published 13 May 2019 Volume 2019:12 Pages 3595—3607
DOI https://doi.org/10.2147/OTT.S184674
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Lu-Jie Miao,1 Shu Yan,2 Qian-Feng Zhuang,3 Qing-Yan Mao,3 Dong Xue,3 Xiao-Zhou He,3 Jian-Ping Chen1
1Department of Gastroenterology, The Third Affiliated Hospital of Soochow University, Changzhou, 213161, People’s Republic of China; 2Department of General Practice, The Third Affiliated Hospital of Soochow University, Changzhou, 213161, People’s Republic of China; 3Department of Urology, The Third Affiliated Hospital of Soochow University, Changzhou, 213161, People’s Republic of China
Background: miR-106b has been reported to play a vital role in pathogenesis of some types of cancer, whilst the role of miR-106b in renal carcinoma cancer (RCC) remains unknown.
Purpose: The objective of this study was to identify the mechanism of miR-106b regulating the progression of renal carcinoma.
Method: The expression of miR-106b was analyzed in RCC cell lines, RCC and adjacent normal renal tissues through qRT-PCR assays. Target mRNA of miR-106b was predicted with databases and verified by luciferase reporter assays. And the effects of miR-106b or targeted mRNA on cell proliferation, invasion, the process of epithelial-mesenchymal transitions (EMTs) were assessed in vitrothrough CCK-8, transwell cell invasion assays, qRT-PCR and Western blotting assays respectively. In addition, the effects of miR-106b on the growth of xenografts mice were analyzedin vivo.
Results: The results demonstrated that miR-106b was significantly increased both in RCC tissues and cell lines. Luciferase reporter assays revealed that miR-106b inhibited Capicua expression by targeting its 3ʹ-UTR sequence. And miR-106b promoted cell proliferation, invasion, EMT progression in RCC cellin vitro, as well as promoted the tumor growth of 786-O cells derived xenografts mice. Additionally, loss of Capicua promoted the activation of MAPK signaling pathway.
Conclusion: The study suggested that miR-106b regulated RCC progression through MAPK signaling pathway partly by targeting Capicua, which might provide valuable evidence for therapeutic target development of RCC.
Keywords: renal carcinoma, miR-106b, capicua, epithelial mesenchymal transitions
Introduction
Renal cell carcinoma cancer (RCC) is a common human kidney cancer, which is the third lethal cancer among the urinary system.1 Approximately one-third patients have metastasis when diagnosed as RCC,2 so it is urgent to find a new molecular biomarker for RCC diagnosis at the early stage. As reported, genetic biomarkers for RCC have been widely investigated including molecules3–5 and microRNAs (miRNAs).6,7
miRNAs is a class of endogenous small non-coding RNAs, the length of which is approximately 19–22 nucleotides. Previous study has reported microRNAs mostly target the 3ʹ- untranslated region and usually repress the gene expression.8 Also miRNAs play important roles in a variety of crucial biological processes including cell apoptosis, growth, invasion and so on.9 And most miRNAs have been identified to participate in the pathogenesis and progression of renal carcinoma cancer, which includes miR-488,10 miR-30a-5p,11 miR-199b-5p7 and so on, which contribute to RCC at different stages through regulating targeted oncogenes or tumor suppressor genes.12 Previous study has reported that miR-106b is involved in the pathological processes of various diseases. Yu et al, found miR-106b enhanced the sensitivity of A549/DDP cells to cisplatin by targeting the expression of PKD2.13 Yao et al, have indicated overexpression of miR-106b shortened the G0/G1 phase and accelerated cell migration and invasion of gastric cancer cells.14 Liu et al, demonstrated that miR-106b negatively regulated osteogenic differentiation of mesenchymal stem cells in vitro.15 However, the role of miR-106b in the progression and development of RCC is still unknown.
Capicua homology protein (CIC) is a member of the Sox–related high-mobility group (HMG) subfamily, which is highly conserved among different species.16 As a transcriptional repressor, Capicua contains two highly conserved domains including HMG-box which mediated DNA binding and nuclear location, and a C-terminal motif C1 which cooperates with the HMG-box for DNA binding.16–19 Recently study has reported that CIC functioned as a transcriptional regulator, and was linked to many cancers and non-cancer diseases including neurodegenerative disease, T cell lymphoblastic leukemia, hepatocellular carcinoma and so on.20–23 Also, several evidences implicated that CIC function as a determinant of sensitivity to EGFR and mitogen-activated protein kinases (MAPK) pathway inhibitors, which suggest that CIC play a broader role in human cancer than originally anticipated. As previous study mentioned, MAPKs are related a family of serine/threonine protein kinase, which is involved in a number of the cellular process including cell proliferation, differentiation and so on.24 However, only a few studies have shown MAPKs participated in the carcinogenesis in RCC,25 and the behavior of which also depends on the different environmental stimuli. Moreover, few studies focused on the regulatory effects of CIC on MAPK signaling in RCC is still unclear. So the present study was aimed to characterize the function of miR-106 in the pathogenesis of RCC and the underlying molecular mechanisms, which might give a new insight into developing a new therapeutic strategy for RCC treatment.
Materials and methods
Cell culture and clinical samples collection
The renal carcinoma cell line 786-O, Caki-1, Caki-2, ACHN and normal renal tubules epithelium cell HK-2 were all purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). HK-2 cells were cultured in keratinocyte serum-free medium supplemented with gentamicin/amphotericin solution (Thermo Fisher Scientific). 786-O, Caki-1, Caki-2 and ACHN cells were all maintained in RPMI-1640 (Hyclone, Logan, MA) medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37°C, the atmosphere contained 5% carbon dioxide (CO2) and 95% air.
A total of 20 fresh paired samples of human primary renal carcinoma and their adjacent normal tissues were collected between 2016 and 2018 at the Third affiliated Hospital of Soochow University (Changzhou, China). Total of 154 RCC pathological sections and clinical characteristics of RCC were collected, and preoperative chemotherapy was not administered to these patients. All patients signed the written informed consent prior to the surgery. The study was approved by the Clinical Research Ethics Committee of the Third affiliated Hospital of Soochow University (Changzhou, China) and was conducted in accordance with the Declaration of Helsinki. The RCC tissues were frozen immediately in liquid nitrogen after collection until use.
Quantitative real-time PCR assays
Total RNA was extracted from tissue samples, and RCC cell lines were exacted using TRIzol reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions.26 Total 2 μg RNA was used to synthesize the first-strand cDNA sequence using the TIANScript II RT kit (TIANGEN, Beijing, China). The relative expression of mRNA was analyzed through qRT-PCR assays with SYBRTM Green ERTM qPCR Supermix (Invitrogen), and β-actin was used as an internal control, the conditions were set as 50 °C for 2 mins, 95 °C 2 mins, 40 cycles with denatured at 95 °C for 15 s, annealed at 60 °C for 15 s, extended at 72 °C for 1 min, and the melting curves were analyzed after 95 °C for 15 s, 60 °C for 15 s, 95 °C for 15 s. The expression of miR-106b from fresh tissues was extracted using TRIzol reagent and detected using miR-XTM miRNA First Strand Synthesis Kit and miR-XTM miRNA qRT-PCR TB GreenTM Kit (TaKaRa, Japan), sncU6 was used as an internal control. The values of miRNA and mRNA expression were both expressed as 2−△△Ct, and sequences of the primers for qRT-PCR are shown in Table 1.
 | Table 1 Primers for qRT-PCR and sequences for CIC siRNA |
To detect the expression of miR-106 in formalin-fixed paraffin-embedded (FFPE) tissues, ~10‑µm‑thick FFPE sections were used. The clinical and pathological characteristics of the patients are shown in Table 2. Total RNA of small interfering RNA FFPE samples was extracted using an miRNeasy FFPE Kit (Qiagen GmbH, Inc.) and detected using miR-XTM miRNA First Strand Synthesis Kit and miR-XTM miRNA qRT-PCR TB GreenTM Kit (Takara, Japan) following the conditions: 95°C for 1 min, followed by 40 cycles at 95°C for 15 s, 55°C for 30 s and 72 °C for 30 s.
 | Table 2 Correlation of miR-106b expression with clinic pathological characteristics of patients |
Plasmid construction and small interfering RNA (siRNA) synthesis
The siRNA for miR-106b (miR-106b inhibitor), miR-106b mimics or negative controls (miR-NC) were all purchased from Gene Pharma Co., Ltd. (Suzhou, China). The lentivirus was prepared following the methods described as previous study,27, and the complementary sequences for miR-106b was cloned into an lv-efla-IRES-EGFP vector by chemical synthesis. Then, the negative control (lv-efla-IRES-EGFP) or miR-106b silencing vector (lv-efla-anti-miR-106b-IRES-EGFP) was co-transfected with D8.91 and pVSV-G into 293T cells. The direct regulatory genes of miR-106b were predicted using PicTar, TargetScan and miRbase Target software. The amplified 3′-UTRs of CIC were cloned into the region directly downstream of a CMV-promoter-driven firefly luciferase cassette in a pmirGO-vector (Promega, Madison, WI, USA). The mutant 3′-UTR of CIC, which carried the mutated sequence in the complementary site was constructed into the pmirGO-wt plasmid.
Cell proliferation assays
Approximately 5,000 cells per well were plated onto 96-well culture plates in 100 µL of RPMI-1640 medium. After transfection with miR-NC, miR-106b mimics, miR-106b inhibitor for 48 hrs, cell viability was assessed using the CCK-8 kit (Beyotime, Shanghai, China). Absorbance was measured at 450 nm using Multiskan FC (Thermo Fisher Scientific, Waltham, MA).
Cell invasion assays
Twenty-four transwell plates (8 μm poresize; BD Biosciences, San Jose, CA) were pre-coated with Matrigel (BD Biosciences) for 30 mins at 37°C. 786-O cells transfected with miR-106b mimics, miR-NC, miR-106b inhibitor were maintained in serum-free medium overnight, and then cells were resuspended at a density of 20,000 cells/mL in medium containing 2% FBS. The cells were then added to the inner chamber of the well, medium containing 20% FBS was added to the outer chamber. After incubation at 37°C for 24 hrs, the inner chamber was fixed with methanol and stained with crystal violet, cells in the inner chamber were removed by cotton swabs, cells on the lower surface of the membrane were counted and photographed.
Luciferase assay
Approximately 5,000 cells per well were plated onto 96-well plates, then cells were co-transfected with 50 nmol/L of miR-106b mimics or miR-NC controls, 50 ng of the luciferase reporter plasmid, and 10 ng of the pRL-CMV Renilla luciferase reporter using 0.5 μL LipofectamineTM 2000 (Invitrogen). After 48 hrs of transfection, cells were seeded and luciferase activity was quantified using a dual-luciferase reporter assay (Promega, Madison, WI, USA).
Western blotting
Transfected 786-O cells were harvested using RIPA lysis Buffer (Beyotime, Shanghai, China) at 4°C for 30 mins and centrifuged at 12,000×g for 5 mins. Concentration of proteins was measured using the BCA assay kit (Biosharp, Anhui, China). Total 40 μg protein samples were separated by 12% SDS-PAGE gel and then transferred into PVDF membranes (Millipore, Billerica, MA), blocked with 5% non-fat milk and incubated overnight with either a rabbit anti-Capicua polyclonal antibody (cat. no. ab123822; dilution, 1:1000; Abcam, Cambridge, MA, US), a rabbit anti-E-cadherin polyclonal antibody (cat. no. ab40772; dilution, 1:25000; Abcam, Cambridge, MA, US), a rabbit anti-N-cadherin polyclonal antibody (cat. no. ab76011; dilution, 1:10000; Abcam, Cambridge, MA), p38 MAPK Rabbit mAb (cat. no. 8690, dilution, 1:1000, Cell Signaling Technology, Danvers, MA), phospho-p38 MAPK (Thr180/Tyr182) Rabbit mAb (cat. no. 4511S, dilution, 1:1000, Cell Signaling Technology, Danvers, MA) or with a mouse anti-β-actin monoclonal antibody (cat. no. EM21002, dilution, 1:6000, Hua An, China). After washed with TBST three times, the membranes were incubated with a goat anti-mouse horseradish peroxides or with goat anti-rabbit horseradish peroxides secondary antibody (Santa Cruz Biotechnology, Dallas, TX) for 2 hrs at room temperature. Expression of proteins was detected using enhanced chemiluminescence reagents (Invitrogen) and X-ray film, the band density was qualified by Image J software, and β-actin was used as internal controls.
In vivo studies
Male Balb/c (nu/nu) mice, 5–6 weeks of age were purchased from the Laboratory Animal Center of Soochow University (Suzhou, China), 1.0×107 786-O cells suspended in 0.1 mL PBS was subcutaneously injected in the right flank of the mice, the protocol is following the previous study. Tumor size was measured by calipers every 5 days. The approximate tumor volume was calculated using the equation V = (length × width × width)/2. Once palpable tumors volumes reached 30–40 mm3, the animals were randomized into three groups. Mice were injected vein tail injection once weekly with anti-miR-106b virus (anti-miR-106b), negative vector virus (NC). When the diameter of tumor reached 15 mm, the mice were sacrificed in accordance with the UK Animals (Scientific Procedures) Act, 1986, and associated guidelines.
Statistical analysis
Results are presented as mean ± standard deviation (sd). miR-106b expression and clinicopathological features were analyzed using Pearson’s chi-squared test. Difference between more than two groups of cell experiments was analyzed using variance (ANOVA) using SPSS 19.0 (Chicago, IL). A value of P<0.05 was considered to be statistically significant.
Results
Expression of mir-106b and CIC in RCC tissues and cell lines
Quantitative real-time PCR analysis revealed that the expression of miR-106b was significantly increased whilst CIC was decreased in RCC tissues compared with adjacent no-tumor renal tissues (P﹤0.05, Figure 1). In addition, the expression levels of miR-106b were also evaluated and CIC were reduced in RCC cell lines compared with the normal human proximal tubule epithelial cell line HK-2. Further, the difference varied among the five RCC cell lines, of which 786-O showed highest miR-106b and lowest CIC expression, so we chose 786-O as the object cell line in the deep study.
Mir-106b serve as a potential independent prognostic marker for RCC
The levels of miR-106b were analyzed with the FFPE renal carcinoma cancer samples. As shown in Table 2, no significant difference was found between miR-106b levels and sex, age, whilst noticeably difference between miR-106 expression and tumor stage, pathology grade and lymph node metastasis. These outcomes suggested that miR-106b may serve as a potential independent prognostic marker for RCC.
Mir-106b promotes proliferation, invasion and EMT in RCC in vitro
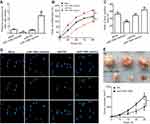
To further investigate whether miR-106b also could affect the progression including proliferation or invasion of RCC cells in vitro, 786-O cells were transfected with miR-106b mimics, miR-NC, miR-106b inhibitor for 48 hrs. CCK-8 and BrdU incorporation assays were used to measure cell proliferative capacity. The results confirmed that the expression of miR-106b was increased in miR-106b mimic groups but decreased in miR-106b inhibitor groups compared with miR-NC groups (Figure 2A). In addition, the proliferation of RCC cells transfected with miR-106b mimics was increased compared with miR-NC groups (Figure 2B–D). In addition, infection of miR-106b silencing virus (anti-miR-106b) decreased the in vivo growth of 786-O cells compared with control (NC). These data further confirmed the effect of miR-106b on RCC cells proliferation (Figure 2E).
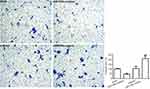
Next, we investigated whether miR-106b could promote renal carcinoma cell invasive processes. As shown in Figure 3, the results revealed that the numbers of invasive 786-O cells with miR-106b mimics were significantly increased compared to those of the miR-NC groups, and the invasive ratio of the cells transfected with miR-106b inhibitor was lower than that transfected with miR-NC.
Since epithelial-mesenchymal transition (EMT) plays a crucial role in the metastasis of various cancer cells including renal carcinoma cancer,28,29 the expressions of EMT-related protein were examined in 786-O cells transfected with target microRNAs. The results demonstrated miR-106b silencing in 786-O cells markedly downregulated the expression of N-cadherin and vimentin, while upregulated the expression of E-cadherin, which lead to the inhibition of EMT (Figure 4).
miR-106b directly targets the 3ʹ-UTR of CIC and suppressed its expression in vitro
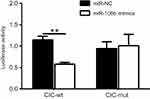
To demonstrate the molecular mechanism by which miR-106b regulates RCC cell proliferation and invasion, bioinformatics databases (PicTar, TargetScan, and PicTar) were used to predict the potential miR-106b targets. The results demonstrated that transfection with miR-106b mimics led to a reduction in the expression of CIC both in mRNA and protein levels (P﹤0.01, Figure 4). Also, the luciferase activity of miR-106b mimics was reduced compared to the control group (P﹤0.01, Figure 5). In addition, when the conserved targeting regions of CIC for miR-106b were mutated, the luciferase activity of the reporter gene was restored. These findings suggested that CIC was a direct target of miR-106b in RCC.
CIC silencing promoted proliferation, invasion of RCC cells in vitro
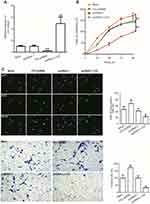
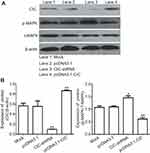
As reported, CIC has been found to be involved in cancer development, whilst the role of CIC in RCC is still unknown. 786-O cells were transfected with CIC silencing vector (CIC-shRNA), CIC overexpressed vector (pcDNA3.1-CIC) or negative control plasmid (pcDNA3.1) for 48 hrs. The results suggested that the expression of CIC was increased in pcDNA3.1-CIC groups but decreased in CIC-shRNA groups compared with negative control groups (Figure 6A). In addition, the proliferation of RCC cells transfected with CIC-shRNA was increased compared with control groups (Figure 6B–6C), and the number of invasive cells of CIC-shRNA groups was increased (Figure 6D). These results suggested that CIC also played a vital role in the progression of 786-O cells in vitro as shown in other cancers.30,31
CIC silencing promoted the activation of the MAPK signaling pathway
To address the mechanism by which CIC regulates RCC, 786-O cells were transfected with CIC-shRNA plasmid or pcDNA3.1-CIC plasmid, the phosphorylation levels of the MAPK signaling pathway were detected. The results showed that the expression of CIC was increased when transfected with CIC overexpression plasmid (pcDNA3.1-CIC) compared with that of cells transfected with pcDNA3.1 control plasmid (pcDNA3.1). Furthermore, phosphorylation levels of MAPK were significantly reduced in pcDNA3.1-CIC groups but increased in CIC-shRNA groups when compared with controls, indicating that CIC silencing activated the MAPK signaling pathway (Figure 7). Collectively, these results suggested that the regulation of renal carcinoma invasion and proliferation by CIC may occur through its activation of the MAPK signaling pathway, in a similar manner to that previously described as BANCR32 and PTEN33 and so on.
Discussion
Renal cell carcinoma is the most lethal type of urologic cancer and accounts for approximately 90% of all renal malignancies worldwide.34 Recently, improved understanding of the molecular mechanism of RCC has received new prospects and more specific focus on the treatment of this disease.35 As reported, a variety of miRNAs function as oncogenic or tumor suppressor in the development and progression of RCC, which provided an available insight into the diagnostic and therapeutic for RCC.36,37 For example, Liu et al, have suggested that miR-142-5p promoted the proliferation and migration of RCC cells by targeting BTG3.38 Perske et al, have shown that RCC cells use miR-146a to reduce or abolish the endogenous NO production to escape macrophage-mediated cell death.39 Lian et al, have proved miR-122 promotes renal carcinoma cancer metastasis through activated PI3K/AKT signaling.40 To our knowledge, this is the first study which demonstrated that miR-106b inhibited RCC growth both in vitro and in vivo by targeting CIC probably through MAPK signaling pathway.
Previously study has reported miR-106b participated the tumorigenesis and progression process in several human tumors, such as non-small cell lung cancer,13 esophageal cancer41 and so on.42 In the current study, we observed that the expression of miR-106b was significantly upregulated in renal carcinoma cancer tissue samples and cell lines when compared with adjacent normal tissues. And miR-106b levels were not correlated with sex, age, whilst noticeable difference was found between miR-106 expression and tumor stage, pathology grade and lymph node metastasis. These outcomes suggested that miR-106b may serve as a potential independent prognostic marker for RCC, which functioned similarly to miR-21 and miR-223,43 miR-154-5p,44 miR-57245 and so on.
Next, we invesitigated the role of miR-106b in the progression of RCC. We found miR-106b silencing suppressed cell proliferation and inhibited the invasive abilities of RCC cells by abrogating EMT (as indicated by upregulation of E-cadherin and downregulation of N-cadherin and vimentin). Since miRNAs exert their biological functions through binding to the 3′-UTR of their target genes, the underlying mechanism by which miR-106b regulated RCC progression was further explored in this study. Through the bioinformatics databases, CIC was selected as one of the possible target gene for miR-106b. As previously mentioned, CIC is downregulated in a variety of human cancers and acted as a tumor suppressor in carcinogenesis.46 In the current study, our results confirmed that miR-106b directly binds to the 3′-UTR of CIC, and overexpression of miR-106b resulted in reduced expressions of CIC at both mRNA and protein levels. Accordingly, these findings implicated that post-transcriptional modulation of CIC by miR-106b might be a pivotal molecular mechanism underlying RCC progression. Notably, an increased expression of miR-106b has been proven to be associated with the reduced sensitivity of cancers to chemotherapeutic agents. Hu et al, suggested miR-106b promoted doxorubicin resistance in breast cancer.47 Mastropasqua et al, showed N-Myc dependent overexpressed miR-106b promoted chemoresistance via regulating of tripartite motif containing 8 (TRIM8) in colorectal cancer.48 On the contrary, demonstrated miR-106b was downregulated in cisplatin-resistance lung cancer cell lines when compared with sensitive parental cells,13 also forced overexpression of miR-106b was associated with increased sensitivity of cancer cells to cisplatin in testicular embryonal carcinoma.49 Collectively, the studies discussed above indicated that miR-106b might have different effects on cellular processes including cellular progression and chemoresistance in various types of human cancer. So in our further study, we are interested in investigating the effects and mechanism of miR-106b participating in chemoresistance in RCC.
The MAPK pathway, known as a cascade of protein kinases, is closely associated with the tumorigenesis in multiple malignancies.50–52 The activation of MAPK pathway could stimulate various oncogenic systems including EMT, which transferred epithelial characteristics into mesenchymal phenotype, finally leading tumors getting migratory and invasive features.53 Consistently, our study also demonstrated that overexpression of CIC inhibited the proliferative and invasion of RCC cells in vivo through suppressing the activation of MAPK pathway, which indicated that MAPK pathway might be involved in the regulation of miR-106b/CIC axis during proliferation and metastasis in RCC progression. To our knowledge, RCC patients who initially experience a clinical benefit with targeted therapy, may acquire alternative pathways to evade therapeutic blockade.54 Buti et al, indicated Everolimus inhibits mTORC1 complex, a downstream effecter of the PI3K/AKT/mTOR pathway involved in cell proliferation and survival.55 Also, mTOR pathway inhibitor temsirolimus is recommended as first-line therapy for metastatic RCC therapy.56 Despite the positive results of these agents reported above in PFS, clinical resistance to mTOR inhibitors usually happened. Work by Ma et al,57 has positioned the RAS/MAPK pathway upstream of mTORC1 was a negative regulator of PI3K signaling. In head and neck squamous cell cancer patients who were non-response to PI3K targeted therapy, mTOR inhibitor in combination with MAPK inhibitor promoted resistance to PI3K inhibition.58 So to this end, miR-106/CIC/MAPK signaling axis may be a promising therapeutic strategy for chemoresistance subset of RCC tumors.
Conclusion
This study implicated the upregulation of miR-106b expression in human RCC tissues and cell lines, and that downregulation of miR-106b could remarkably inhibit cell proliferation, invasion, EMT in vitro by targeting CIC. Furthermore, forced CIC expression suppressed the activation of the MAPK signaling pathway. Therefore, these data suggested that miR-106b functioned as a novel oncogene that regulated RCC progression through the MAPK signaling pathway by targeting CIC, which might provide valuable evidence for therapeutic target development for RCC.
Acknowledgment
This study was supported by grants from the Funding for National Science Foundation of Jiangsu Province (Grant no. BK20150251), Youth Medical Talent Project of Jiangsu Province (QNRC2016292), China Postdoctoral Science Foundation (Grant no 63, 2018M632371).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Siska PJ, Beckermann KE, Rathmell WK, Haake SM. Strategies to overcome therapeutic resistance in renal cell carcinoma. Urol Oncol. 2017;35(3):102–110. doi:10.1016/j.urolonc.2016.12.002
3. Wu YY, Liu XY, Zhuo DX, Huang HB, Zhang FB, Liao SF. Decreased expression of TRPV1 in renal cell carcinoma: association with tumor Fuhrman grades and histopathological subtypes. Cancer Manag Res. 2018;10:1647–1655. doi:10.2147/CMAR.S166390
4. Lee HJ, Shin DH, Kim SY, et al. TFE3 translocation and protein expression in renal cell carcinoma are correlated with poor prognosis. Histopathology. 2018. doi:10.1111/his.13700
5. Chen D, Chen W, Xu Y, et al. Upregulated immune checkpoint HHLA2 in clear cell renal cell carcinoma: a novel prognostic biomarker and potential therapeutic target. J Med Genet. 2019;56(1):43–49. doi:10.1136/jmedgenet-2018-105454.
6. Yamada Y, Arai T, Kojima S, et al. Regulation of anti-tumor miR-144-5p targets oncogenes: direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018. doi:10.1111/cas.13722
7. Lai Y, Quan J, Hu J, et al. miR-199b-5p serves as a tumor suppressor in renal cell carcinoma. Exp Ther Med. 2018;16(1):436–444. doi:10.3892/etm.2018.6151
8. Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172(3):962–974. doi:10.1016/j.jconrel.2013.09.015
9. Ciesla M, Skrzypek K, Kozakowska M, Loboda A, Jozkowicz A, Dulak J. MicroRNAs as biomarkers of disease onset. Anal Bioanal Chem. 2011;401(7):2051–2061. doi:10.1007/s00216-011-5001-8
10. Wei X, Yu L, Kong X. miR-488 inhibits cell growth and metastasis in renal cell carcinoma by targeting HMGN5. Onco Targets Ther. 2018;11:2205–2216. doi:10.2147/OTT.S156361
11. Liu E, Sun X, Li J, Zhang C. miR30a5p inhibits the proliferation, migration and invasion of melanoma cells by targeting SOX4. Mol Med Rep. 2018;18(2):2492–2498.doi:10.3892/mmr.2018.9166.
12. Li M, Wang Y, Song Y, et al. MicroRNAs in renal cell carcinoma: a systematic review of clinical implications (Review). Oncol Rep. 2015;33(4):1571–1578. doi:10.3892/or.2015.3799
13. Yu S, Qin X, Chen T, Zhou L, Xu X, Feng J. MicroRNA-106b-5p regulates cisplatin chemosensitivity by targeting polycystic kidney disease-2 in non-small-cell lung cancer. Anticancer Drugs. 2017;28(8):852–860. doi:10.1097/CAD.0000000000000524
14. Yao YL, Wu XY, Wu JH, et al. Effects of microRNA-106 on proliferation of gastric cancer cell through regulating p21 and E2F5. Asian Pac J Cancer Prev. 2013;14(5):2839–2843.
15. Liu K, Jing Y, Zhang W, et al. Silencing miR-106b accelerates osteogenesis of mesenchymal stem cells and rescues against glucocorticoid-induced osteoporosis by targeting BMP2. Bone. 2017;97:130–138. doi:10.1016/j.bone.2017.01.014
16. Lee CJ, Chan WI, Cheung M, et al. CIC, a member of a novel subfamily of the HMG-box superfamily, is transiently expressed in developing granule neurons. Brain Res Mol. 2002;106(1–2):151–156.
17. Jimenez G, Guichet A, Ephrussi A, Casanova J. Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes Dev. 2000;14(2):224–231.
18. Astigarraga S, Grossman R, Diaz-Delfin J, Caelles C, Paroush Z, Jimenez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. Embo J. 2007;26(3):668–677. doi:10.1038/sj.emboj.7601532
19. Fores M, Simon-Carrasco L, Ajuria L, et al. A new mode of DNA binding distinguishes Capicua from other HMG-box factors and explains its mutation patterns in cancer. PLoS Genet. 2017;13(3):e1006622. doi:10.1371/journal.pgen.1006622
20. Tan Q, Brunetti L, Rousseaux MWC, et al. Loss of Capicua alters early T cell development and predisposes mice to T cell lymphoblastic leukemia/lymphoma. Proc Natl Acad Sci U S A. 2018;115(7):E1511–E9. doi:10.1073/pnas.1716452115
21. Kim E, Kim D, Lee JS, et al. Capicua suppresses hepatocellular carcinoma progression by controlling the ETV4-MMP1 axis. Hepatology. 2018;67(6):2287–2301. doi:10.1002/hep.29738
22. Kawamura-Saito M, Yamazaki Y, Kaneko K, et al. Fusion between CIC and DUX4 up-regulates PEA3 family genes in Ewing-like sarcomas with t(4;19)(q35;q13) translocation. Hum Mol Genet. 2006;15(13):2125–2137. doi:10.1093/hmg/ddl136
23. Lam YC, Bowman AB, Jafar-Nejad P, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–1347. doi:10.1016/j.cell.2006.11.038
24. Dunn KL, Espino PS, Drobic B, He S, Davie JR. The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem Cell Biol. 2005;83(1):1–14. doi:10.1139/o04-121
25. Oka H, Chatani Y, Hoshino R, et al. Constitutive activation of mitogen-activated protein (MAP) kinases in human renal cell carcinoma. Cancer Res. 1995;55(18):4182–4187.
26. Kotlarek M, Kubiak A, Jazdzewski K, Wojcicka A. MicroRNA analysis using the quantitative real-time PCR reaction. Methods Mol Biol. 2018;1823:69–85. doi:10.1007/978-1-4939-8624-8_7
27. Yang H, Song E, Shen G, et al. Expression of microRNA-30c via lentivirus vector inhibits the proliferation and enhances the sensitivity of highly aggressive ccRCC Caki-1 cells to anticancer agents. Onco Targets Ther. 2017;10:579–590. doi:10.2147/OTT.S115791
28. Kaur G, Li CG, Chantry A, Stayner C, Horsfield J, Eccles MR. SMAD proteins directly suppress PAX2 transcription downstream of transforming growth factor-beta 1 (TGF-beta1) signalling in renal cell carcinoma. Oncotarget. 2018;9(42):26852–26867. doi:10.18632/oncotarget.25516
29. Zhao S, Wang Y, Luo M, Cui W, Zhou X, Miao L. Long noncoding rna small nucleolar rna host gene 1 (SNHG1) promotes renal cell carcinoma progression and metastasis by negatively regulating miR-137. Med Sci Monit. 2018;Jun(24):3824–3831. doi:10.12659/MSM.910866
30. Chebib I, Jo VY. Round cell sarcoma with CIC-DUX4 gene fusion: discussion of the distinctive cytomorphologic, immunohistochemical, and molecular features in the differential diagnosis of round cell tumors. Cancer Cytopathol. 2016;124(5):350–361. doi:10.1002/cncy.21685
31. Yang R, Chen LH, Hansen LJ, et al. Cic loss promotes gliomagenesis via aberrant neural stem cell proliferation and differentiation. Cancer Res. 2017;77(22):6097–6108. doi:10.1158/0008-5472.CAN-17-1018
32. Zhang J, Du Y, Zhang X, Li M, Li X. Downregulation of BANCR promotes aggressiveness in papillary thyroid cancer via the MAPK and PI3K Pathways. J Cancer. 2018;9(7):1318–1328. doi:10.7150/jca.20150
33. Carabia J, Carpio C, Abrisqueta P, et al. Microenvironment regulates the expression of miR-21 and tumor suppressor genes PTEN, PIAS3 and PDCD4 through ZAP-70 in chronic lymphocytic leukemia. Sci Rep. 2017;7(1):12262. doi:10.1038/s41598-017-12135-7
34. Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): a literature review. Cancer Treat Rev. 2008;34(3):193–205. doi:10.1016/j.ctrv.2007.12.001
35. Pal SK, Nelson RA, Vogelzang N. Disease-specific survival in de novo metastatic renal cell carcinoma in the cytokine and targeted therapy era. PLoS One. 2013;8(5):e63341. doi:10.1371/journal.pone.0063341
36. Wei S, Bing Z, Yao Y, Master SR, Gupta P. Higher expression of mir-182 in cytology specimens of high-grade urothelial cell carcinoma: a potential diagnostic marker. Acta Cytol. 2015;59(1):109–112. doi:10.1159/000371507
37. Khella HW, Scorilas A, Mozes R, et al. Low expression of miR-126 is a prognostic marker for metastatic clear cell renal cell carcinoma. Am J Pathol. 2015;185(3):693–703. doi:10.1016/j.ajpath.2014.11.017
38. Liu L, Liu S, Duan Q, et al. MicroRNA-142-5p promotes cell growth and migration in renal cell carcinoma by targeting BTG3. Am J Transl Res. 2017;9(5):2394–2402.
39. Perske C, Lahat N, Sheffy Levin S, Bitterman H, Hemmerlein B, Rahat MA. Loss of inducible nitric oxide synthase expression in the mouse renal cell carcinoma cell line RENCA is mediated by microRNA miR-146a. Am J Pathol. 2010;177(4):2046–2054. doi:10.2353/ajpath.2010.091111
40. Lian JH, Wang WH, Wang JQ, Zhang YH, Li Y. MicroRNA-122 promotes proliferation, invasion and migration of renal cell carcinoma cells through the PI3K/Akt signaling pathway. Asian Pac J Cancer Prev. 2013;14(9):5017–5021.
41. Gu J, Wang Y, Wu X. MicroRNA in the pathogenesis and prognosis of esophageal cancer. Curr Pharm Des. 2013;19(7):1292–1300.
42. He QY, Wang GC, Zhang H, et al. miR-106a-5p suppresses the proliferation, migration, and invasion of osteosarcoma cells by targeting HMGA2. DNA Cell Biol. 2016;35(9):506–520. doi:10.1089/dna.2015.3121
43. Yu L, Xiang L, Feng J, et al. miRNA-21 and miRNA-223 expression signature as a predictor for lymph node metastasis, distant metastasis and survival in kidney renal clear cell carcinoma. J Cancer. 2018;9(20):3651–3659.
44. Lin C, Li Z, Chen P, et al. Oncogene miR-154-5p regulates cellular function and acts as a molecular marker with poor prognosis in renal cell carcinoma. Life Sci. 2018;15(209):481–489. doi:10.1016/j.lfs.2018.08.044
45. Pan X, Li Z, Zhao L, et al. microRNA‑572 functions as an oncogene and a potential biomarker for renal cell carcinoma prognosis. Oncol Rep. 2018;40(5):3092–3101. doi:10.3892/or.2018.6649
46. Simon-Carrasco L, Jimenez G, Barbacid M, Drosten M. The Capicua tumor suppressor: a gatekeeper of Ras signaling in development and cancer. Cell Cycle. 2018;17(6):702–711. doi:10.1080/15384101.2018.1450029
47. Hu Y, Li K, Asaduzzaman M, et al. MiR-106b~25 cluster regulates multidrug resistance in an ABC transporter-independent manner via downregulation of EP300. Oncol Rep. 2016;35(2):1170–1178. doi:10.3892/or.2015.4412
48. Mastropasqua F, Marzano F, Valletti A, et al. TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours. Mol Cancer. 2017;16(1):67. doi:10.1186/s12943-017-0634-7
49. Koster R, Di Pietro A, Timmer-Bosscha H, et al. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J Clin Invest. 2010;120(10):3594–3605. doi:10.1172/JCI41939
50. Cai Y, Dong ZY, Wang JY. LncRNA NNT-AS1 is a major mediator of cisplatin chemoresistance in non-small cell lung cancer through MAPK/Slug pathway. Eur Rev Med Pharmacol Sci. 2018;22(15):4879–4887. doi:10.26355/eurrev_201808_15624
51. Chen Q, Li KT, Tian S, et al. Photodynamic therapy mediated by aloe-emodin inhibited angiogenesis and cell metastasis through activating MAPK signaling pathway on HUVECs. Technol Cancer Res Treat. 2018;Jan(17):1533033818785512.
52. Sheng L, Tang T, Liu Y, et al. Inducible HSP70 antagonizes cisplatininduced cell apoptosis through inhibition of the MAPK signaling pathway in HGC27 cells. Int J Mol Med. 2018;42(4):2089–2097. doi:10.3892/ijmm.2018.3789
53. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5(5):402–418. doi:10.1016/j.apsb.2015.07.005
54. De Lisi D, De Giorgi U, Lolli C, et al. Lenvatinib in the management of metastatic renal cell carcinoma: a promising combination therapy? Expert Opin Drug Metab Toxicol. 2018;14(4):461–467. doi:10.1080/17425255.2018.1455826
55. Buti S, Leonetti A, Dallatomasina A, Bersanelli M. Everolimus in the management of metastatic renal cell carcinoma: an evidence-based review of its place in therapy. Core Evid. 2016;11:23–36. doi:10.2147/CE.S98687
56. Hudes G, Carducci M, Tomczak P, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356(22):2271–2281. doi:10.1056/NEJMoa066838
57. Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121(2):179–193. doi:10.1016/j.cell.2005.02.031
58. Ruicci KM, Pinto N, Khan MI, et al. ERK-TSC2 signalling in constitutively-active HRAS mutant HNSCC cells promotes resistance to PI3K inhibition. Oral Oncol. 2018;84:95–103. doi:10.1016/j.oraloncology.2018.07.010
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.