Back to Journals » Breast Cancer: Targets and Therapy » Volume 14
miR-101-5p Acts as a Tumor Suppressor in HER2-Positive Breast Cancer Cells and Improves Targeted Therapy
Authors Normann LS , Haugen MH, Aure MR, Kristensen VN, Mælandsmo GM, Sahlberg KK
Received 8 September 2021
Accepted for publication 2 February 2022
Published 1 March 2022 Volume 2022:14 Pages 25—39
DOI https://doi.org/10.2147/BCTT.S338404
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Lisa Svartdal Normann,1– 3 Mads Haugland Haugen,2 Miriam Ragle Aure,3 Vessela N Kristensen,3 Gunhild Mari Mælandsmo,2,4 Kristine Kleivi Sahlberg1,2
1Department of Research and Innovation, Vestre Viken Hospital Trust, Drammen, Norway; 2Department of Tumor Biology, Institute for Cancer Research, Oslo University Hospital, Oslo, Norway; 3Department of Medical Genetics, Institute of Clinical Medicine, Faculty of Medicine, University of Oslo and Oslo University Hospital, Oslo, Norway; 4Institute for Medical Biology, Faculty of Health Sciences, UiT – The Arctic University of Norway, Tromsø, Norway
Correspondence: Kristine Kleivi Sahlberg, Department of Research and Innovation, Vestre Viken Hospital Trust, Drammen, Norway, Tel +47 98641229, Email [email protected]
Purpose: Human epidermal growth factor receptor 2 positive (HER2+) breast cancers responding poorly to targeted therapy need improved treatment options. miR-101-5p has shown tumor-suppressive properties in multiple cancer forms, and we assessed the effect and mechanism of action of this miRNA in HER2+ breast cancer.
Methods: Expression levels of miR-101-5p in two clinical datasets, TCGA and METABRIC, were compared between tumor and normal adjacent samples, and across molecular subtypes and HER2 status. The ability of miR-101-5p to sensitize for treatment with lapatinib, tucatinib and trastuzumab was explored in HER2+ breast cancer cells responding poorly to such targeted drugs. Proliferation and apoptosis assays and downstream protein analysis were performed.
Results: Expression levels of miR-101-5p were significantly lower in tumor compared to normal adjacent tissue (p < 0.001), and particularly low in HER2+ tumors, both the HER2-enriched subtype (p ≤ 0.037) and clinical HER2-status (p < 0.001). In a HER2+ cell line (KPL4) responding poorly to targeted drugs, miR-101-5p overexpression inhibited proliferation (p < 0.001), and combinatorial treatment with lapatinib and trastuzumab significantly further decreased this inhibition (p = 0.004). Proteomic data and in silico analyses revealed PI3K/Akt- and HER2-pathways among the predicted regulated pathways. miR-101-5p alone (p = 0.018) and in combination with lapatinib and trastuzumab (p < 0.001) induced apoptosis, while the targeted drugs alone did not exert any significant effect neither on proliferation nor apoptosis.
Conclusion: miR-101-5p acts as a tumor suppressor by inducing apoptosis in HER2+ breast cancer and sensitizes cells with initially poor response to lapatinib and trastuzumab.
Keywords: microRNA, targeted treatment, apoptosis, sensitize
Introduction
Human epidermal growth factor receptor 2 positive (HER2+) breast cancer is an aggressive disease that accounts for about 15% of all breast cancer cases.1,2 HER2+ breast cancer cells have an overexpression of the tyrosine kinase HER2 on their surface, which initiates signaling pathways that regulate cellular growth, proliferation and survival. Based on a breast tumor’s expression of hormone-receptors, HER2 and Ki-67 protein levels, they are categorized as either luminal A, luminal B, triple-negative or HER2-enriched. The latter shows the highest rate of local recurrence and distant metastasis, in addition to the lowest 10-year recurrence free survival.3 There are multiple drugs that specifically target and inhibit HER2 in clinical use, including monoclonal antibodies (trastuzumab, pertuzumab, margetuximab), small molecular kinase inhibitors (lapatinib, neratinib, tucatinib) and monoclonal antibodies linked to chemotherapy (trastuzumab emtansine, fam-trastuzumab deruxtecan-nxki). It is expected that current recurrence rates will decrease in follow-up studies where larger portions of the patients have received targeted therapy.3 However, despite the available targeted therapies, some patients still experience disease progression upon primary or acquired resistance. The majority of patients with HER2+ metastatic breast cancer die from their disease,4–6 and a continuous search for improved treatment options is needed.
MicroRNAs (miRNAs) are short (18–24 nucleotides in length), single-stranded, non-coding, post-transcriptional regulators of cellular processes.7 They are endogenously expressed in cells and regulate gene expression by binding to complementary messenger RNA (mRNA) strands. This binding leads to mRNA degradation or prevention of translation, and thus affects the proteome and is considered to participate in regulation of numerous biological processes in the cell.
During the synthesis of miRNA, nucleotide duplexes with a 3’-end (−3p) and 5’-end (−5p) are cleaved into single strands. Historically, due to differences in mature expression level, the −5p strand was considered the guide strand with the dominant biological activity, while the −3p strand was considered the passenger strand. However, it has become evident that both strands function as post-transcriptional regulators.8,9 There is a substantial amount of research performed on hsa-miR-101-3p, both regarding its intrinsic low levels in multiple cancer forms and its effect on cancer cell viability, migration and invasion.10–14 It is, however, far less knowledge on hsa-miR-101-5p (miR-101-5p), but interestingly, Toda et al recently published a study depicting the tumor- suppressive properties of miR-101-5p in breast cancer.15 This included the relationship between high miR-101-5p expression and improved overall survival, and the identification of seven putative oncogenic gene targets.
There are currently (January 2022) 1095 studies registered on ClinicalTrials.gov including the term “microRNA”, of which 52 are related to breast cancer. These trials are all focused on the use of miRNAs as biomarkers, but there is also an emerging focus on miRNAs as therapeutic agents.16 Several miRNA therapeutics have been through Phase 1 and 2 clinical trials, including the miR-122 repressor Miravirsen for hepatitis C virus treatment,17,18 and bacterially derived minicells (TargomiRs) loaded with miR-16 for recurrent malignant pleural mesothelioma.19 The latter example involves nanoparticles directed at the epidermal growth factor receptor (EGFR), which mesothelioma cells frequently overexpress on their surface. Such a delivery method is of particular interest for HER2 overexpressing breast cancer cells and lays a promising ground for future miRNA therapeutics.
We have previously shown the promising effects of combining miR-101-5p overexpression with trastuzumab and lapatinib to inhibit cell viability in HER2+ breast cancer cells that respond poorly to targeted treatment.20 A high-throughput miRNA screen revealed miR-101-5p as one of eight promising miRNAs with sensitizing properties in HER2+ breast cancer cells. In the current study, we aim to investigate the molecular mechanisms that cause the sensitizing effect of miR-101-5p by studying changes in the proteomic landscape in HER2 cancer cells upon treatment.
Materials and Methods
Clinical Data
miRNA expression, breast cancer subtype and HER2 status data from two publicly available breast cancer patient cohorts, The Cancer Genome Atlas Network (TCGA; n = 825 patients)21 and The Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; n = 1394 patients),22 were used for expression analyses. Claudin-low subtype was only available for the METABRIC dataset. For TCGA, miRNA expression data in the form of log2 (RPM + 1) miRNA mature strand expression (level 3) measured by Illumina HiSeq, together with clinical data were downloaded from the UCSC Xena browser.23 For the METABRIC cohort, normalized miRNA expression data measured by Agilent ncRNA 60k arrays were downloaded from The European Genome-phenome Archive (EGA), accession number EGAD00010000438. Ethical approval for research on human data were given from the Regional Committees for Medical Research Ethics South East Norway – Faculty of Medicine, Committee C (project approval number: 2016/433). Statistical significance between groups was calculated using Wilcoxon rank-sum tests.
Cell Lines
KPL4 cells were kindly provided by Professor J. Kurebayashi (Kawasaki Medical School, Japan).24 JIMT1 cells were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ; Braunschweig, Germany). KPL4 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; 4.5 g/L glucose) supplemented with 10% fetal bovine serum, 1% Glutamax and 2% HEPES (Sigma-Aldrich, St. Louis, MO, USA). JIMT1 cells were cultured in 1:1 DMEM and Ham’s F-12 (Corning Inc., Corning, NY, USA) supplemented with 10% fetal bovine serum and 1% Glutamax. The cells were incubated at 37 °C, 5% CO2, and used for less than 30 passages. Both cell lines were chosen based on their poor response to the HER2-targeted therapies trastuzumab and lapatinib.25
miRNA Transfection and Treatment with HER2-Targeting Drugs
KPL4 and JIMT1 cells were transfected with miR-101-5p mimic and miRIDIAN Mimic Negative Control #1 (Dharmacon, Lafayette, CO, USA) at a final concentration of 20 nM. SiLentFect Lipid Reagent (Bio-Rad Laboratories, Hercules, CA, USA) was diluted in Opti-MEM Reduced Serum Medium, no phenol red (Thermo Fisher Scientific, Waltham, MA USA), and incubated at room temperature for 10 minutes, then mixed with either miR-101-5p or the scrambled negative control. The miRNA/lipid solutions were incubated for 1 hour at room temperature. The cells were transfected in 96-well plates (10000 cells; Costar 96-well white, clear-bottom polystyrene plates, Corning Inc., Corning, NY, USA), and confluence was measured using IncuCyte FLR live cell imaging (Essen BioScience, Ann Arbor, MI, USA). For protein analysis, KPL4 cells were transfected in T25 flasks (1.2 million cells; Thermo Fisher Scientific) for 48 hours. The transfected cells were simultaneously treated with 100 nM lapatinib ditosylate (GW-572016, Selleckchem, Houston, TX, USA), or 2.5 µM tucatinib (ONT-380, Selleckchem), in combination with 10 µg/mL trastuzumab (Roche Applied Biosciences, Basel, Switzerland). Statistical difference between treatment groups was assessed with Student’s t-tests.
Protein Lysates
T25 flasks with untreated/treated (trastuzumab and lapatinib) transfected KPL4 cells were washed twice with phosphate-buffered saline (PBS; Sigma-Aldrich) and the cells were harvested using a cell scraper (no. 83.1832, Sarstedt AG, Nümbrecht, Germany). The cells were transferred to a 15-mL tube (Sarstedt AG) and centrifuged. The supernatant was discarded, and the cell pellet was covered in M-PER lysis buffer (Thermo Fisher Scientific) containing PhosStop Phosphatase Inhibitor Cocktail Tablets and cOmplete Protease Inhibitor Cocktail Tablets (Roche Applied Biosciences). The samples were incubated on ice for 20 minutes, with vortexing every 5 minutes. Next, the samples were centrifuged at 13,000 rpm for 15 minutes at 4 °C, and the supernatant protein lysate collected. Total protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol, with readout on a Victor X Plate Reader (PerkinElmer, Norwalk, CT, USA).
Reverse Phase Protein Array (RPPA)
Protein lysates were further prepared according to instructions from MD Anderson for use on their RPPA platform. The RPPA method26 in short included serial dilutions of the lysates before printing them on nitrocellulose-coated slides. The slides were probed with 454 primary antibodies and appropriate biotinylated secondary antibodies. A streptavidin-conjugated HRP and DAB colorimetric reaction was used for signal visualization. The slides were scanned, and spot intensities were measured. Relative protein levels were estimated using a fitted curve (“supercurve”), and subsequently normalized for protein loading. Data was analyzed in R (version 4.0.2; R Foundation for Statistical Computing, Vienna, Austria) in R Studio (version 1.1.423).27 Heatmaps with Pearson average clustering were prepared using the R package clustermap (version 1.2.3)28 using log2-transformed and median centered data with tanh adjustment for visualization to compress the extreme values.
In silico Pathway Analyses
Ingenuity Pathway Analyses (IPA) software (version 57662101; QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis)29 was used for in silico pathway analyses, and to determine a list of HER2 pathway-related genes. The Enrichr online tool was used for pathway enrichment analyses,30,31 where Human KEGG 2019 pathway data were downloaded on May 2, 2021.
Apoptosis Assay
Induction of apoptosis upon miRNA transfection and/or drug treatment with trastuzumab and lapatinib was assessed using IncuCyte Caspase-3/7 Red Dye (Essen BioScience). KPL4 cells were seeded in Costar 96 well-plates (20,000 cells/well; Corning Inc.) and transfected with miR-101-5p or miRIDIAN Mimic Negative Control #1 (20 nM; Dharmacon) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific). The cells were simultaneously treated with lapatinib (100 nM; SelleckChem) and trastuzumab (10 µg/mL; Roche Applied Biosciences). Caspase-3/7 Red Dye was added, and fluorescence intensity was measured and analyzed using the IncuZoom live-cell analysis microscope and software (Essen BioScience) for 48 hours. Statistical difference at the time endpoint was calculated using Student’s t-test.
Results
miR-101-5p is Downregulated in Breast Cancer and Particularly in HER2+ Patients
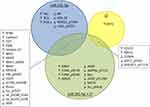
To assess the expression of miR-101-5p in breast cancer tissue compared to normal adjacent tissue, data from the TCGA breast cancer patient cohort was used. This demonstrated that miR-101-5p was significantly downregulated in cancer tissue (Wilcoxon rank-sum test, p < 0.001; Figure 1A), and confirms a previous finding from an independent study using a smaller dataset (n = 27).15 The same trend was observed in the METABRIC dataset, without reaching the p < 0.05 level of statistical significance (Wilcoxon rank-sum test, p = 0.069; Figure 1B). Interestingly, when stratifying the breast cancer patients into molecular subtypes and sorting the groups based on miR-101-5p expression, the HER2-enriched subtype was the group with the lowest expression level in both patient datasets (Figure 1C and D). Of note, there was not a significant difference between the basal and the HER2-enriched subtypes in METABRIC (Wilcoxon rank-sum test, p = 0.788; Figure 1D). However, when using the immunohistochemical classification of HER2 status, the level of miR-101-5p was significantly lower in HER2+ breast cancer patients than in HER2- patients, both in the TCGA and the METABRIC cohorts (Wilcoxon rank-sum test p < 0.001 in both cohorts; Figure 1E and F).
miR-101-5p Sensitizes for Targeted Treatment in HER2+ Cells
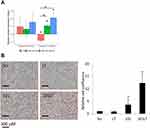
We have previously shown that overexpression of miR-101-5p reduces cell viability measured by CellTiter-Glo assay in HER2+ KPL4 breast cancer cells alone and in combination with lapatinib and trastuzumab.20 These data were confirmed here using a complementary cell proliferation assay measuring the confluence over time after miR-101-5p overexpression. Indeed, miR-101-5p overexpression inhibited proliferation compared to the scrambled negative control (Student’s t-test, p < 0.001; Figure 2A). The addition of lapatinib and trastuzumab to the miRNA overexpression further decreased cell proliferation compared to the miRNA alone (Student’s t-test p = 0.004). Importantly, a significant downregulation of cell proliferation by lapatinib and trastuzumab was not observed compared to the negative control (Student’s t-test, p = 0.166). Similarly, the combination of trastuzumab and tucatinib did not significantly downregulate cell proliferation (Figure 2B, Student’s t-test, p = 0.152). When combining trastuzumab and tucatinib with miR-101-5p, there was a trend towards improved inhibition of cell proliferation, although not significant compared to the miRNA alone (Student’s t-test, p = 0.298). In JIMT1 cells, miR-101-5p significantly inhibited cell proliferation (Student’s t-test, p = 0.005), but the addition of lapatinib and trastuzumab or tucatinib and trastuzumab did not significantly enhance this inhibition (Figure 2C and D). The proliferation inhibition by the miRNA alone proposes a tumor suppressor effect of miR-101-5p in two HER2+ cell lines. Furthermore, the results suggest the potential value of a combinatorial treatment using this miRNA together with lapatinib and trastuzumab in selected patient groups.
Combinatorial Treatment Affects Signaling Pathways Involved in Proliferation and Survival
To investigate mechanisms involved in growth inhibition by miR-101-5p and the targeted drugs in KPL4 cells, we studied changes in the proteomic landscape using the RPPA method. Unsupervised clustering of the protein data showed clear distinction between cells overexpressing miR-101-5p and cells transfected with a scrambled negative control (Figure 3A). This indicates a consistent global change in protein expression after miR-101-5p transfection.
 |
Figure 3 Changes in the proteomic landscape after transfection with miR-101-5p (101) or scrambled negative control (Scr) and treatment with lapatinib and trastuzumab (LT). (A) Protein lysates analyzed by an RPPA assay of 454 protein antibodies presented as a heatmap. Unsupervised Pearson average clustering of the 12 samples with dendrograms illustrating similarities between samples/proteins. Unsupervised Pearson average clustering resulted in two distinct annotation clusters for the 101 and the Scr transfected KPL4 cells. Five distinct clusters between proteins were obtained. (B) The proteins from the bottom, dark blue cluster (cluster five; n = 71 proteins) were subjected to Enrichr online pathway analysis,30,31 and the top eight enriched pathways are presented. |
The 454 analyzed proteins formed five clusters with Pearson average unsupervised clustering, and the mean log2 protein levels within each cluster were calculated separately for the miR-101-5p and the scrambled negative control transfected cells (Figure 3A; Supplementary Table S1). The bottom dark blue cluster showed a strong downregulation of the proteins in the miR-101-5p overexpressing samples (mean log2 expression = −0.19 compared to mean log2 expression = 0.07 for the scrambled transfected cells; Supplementary Table S1). The proteins in this cluster were subjected to in silico pathway enrichment analyses and were predicted to be involved in proliferation, survival and cancer signaling pathways, such as PI3K-Akt, mTOR and ErbB signaling (Figure 3B). This suggests that miR-101-5p alone or in combination with targeted treatment affects these pathways.
To classify the effects on expression of single proteins after treatment with miR-101-5p and the combination of lapatinib and trastuzumab, we set a cut-off for significant alterations at a fold-change (FC) > |0.4| and statistical significance (Student’s t-test, unadjusted for multiple testing) p < 0.05 compared to the control. This resulted in 25 significantly altered proteins for miR-101-5p (Table 1), six proteins for lapatinib and trastuzumab (Table 2) and 33 proteins for the combination of miR-101-5p and trastuzumab and lapatinib (Table 3). All protein alterations are presented in Supplementary Table S2. Multiple proteins overlapped between the different treatment groups (Figure 4), but surprisingly no protein overlapped all three comparisons. The relative protein expressions upon the different treatments are visualized for the 33 proteins significantly altered with the combinatorial treatment (Figure 5). The distinct downregulation of HER3, Rictor, and phosphorylated mTOR, S6 and Src are among the protein alterations that may explain the negative effect on cell proliferation.32–34 Overall, the observed fold change values were greater in the combination group compared to the mono-treatment groups.
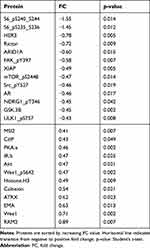 |
Table 1 Protein Alterations in KPL4 Cells Transfected with miR-101-5p Compared to Scrambled Negative Control |
 |
Table 2 Protein Alterations in KPL4 Cells Treated with Lapatinib and Trastuzumab Compared to Scrambled Negative Control |
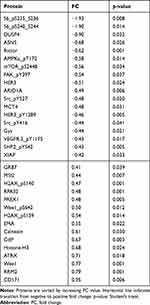 |
Table 3 Protein Alterations in KPL4 Cells Transfected with miR-101-5p and Treated with Lapatinib and Trastuzumab Compared to Scrambled Negative Control |
Effect on HER2 Signaling Pathway
Focusing on HER2+ breast cancer patients, we studied the impact on the proteins in the HER2 axis. We used in silico analysis to generate a list of HER2 pathway relevant genes (n = 76, Supplementary Table S3) and compared this to the genes corresponding to the proteins, which showed significant changes in expression upon treatment. Of the proteins significantly altered in expression by miR-101-5p in combination with lapatinib and trastuzumab (Figure 5), five proteins were central in the HER2 pathway: HER3, mTOR, S6, SRC and XIAP, all of which were downregulated (Table 3). From the lapatinib and trastuzumab altered proteins, only one, EGFR, was related to the HER2 pathway. This cell line is known to respond poorly to HER2 targeted treatment,25 which may explain why so few HER2 pathway-related proteins were altered. Using in silico analysis, fold change values of the significantly altered proteins after the combination treatment were projected onto the HER2 signaling pathway (Supplementary Figure S1). Analysis of protein regulation predicted, eg an upregulation of apoptosis and a downregulation of cell survival, progression, metastasis, invasion and epithelial-to-mesenchymal transition. Furthermore, the ten proteins exclusively regulated by the combinatorial treatment (Figure 4) were subjected to pathway analysis and HER2-signaling in breast cancer was among the top 10 relevant canonical pathways (Supplementary Table S3).
miR-101-5p Sensitize Cells to Targeted Treatment and Induce Apoptosis
Based on the in silico predicted upregulation of apoptosis, we studied the specific alteration of two apoptosis-related proteins upon treatment: cleaved caspase 3 and 7. All treatments showed a tendency towards increased levels of cleaved caspase 3 (Figure 6A), but none were significant. Surprisingly, lapatinib and trastuzumab alone decreased the level of cleaved caspase 7 significantly (Figure 6A; Student’s t-test, p = 0.046). Treatment with miR-101-5p overexpression significantly increased this level, both compared to the targeted drugs and scrambled negative control (Scr; p = 0.014 and p = 0.019, respectively). Interestingly, the level of cleaved caspase 7 increased further in the miR-101-5p and lapatinib and trastuzumab combination treatment, although not significant from the miR-101-5p treatment alone (p = 0.149). Other proteins suggesting apoptosis were, eg decreased measurements of the anti-apoptotic regulators Mcl-1, XIAP and Bcl-xL (the latter increased with miR-101-5p, and significantly decreased with the addition of lapatinib and trastuzumab; Supplementary Figure S2).
This trend towards increased apoptosis was of interest to investigate further. Using a live-cell imaging apoptosis assay, we observed that overexpression of miR-101-5p significantly induced apoptosis compared to scrambled negative control and lapatinib and trastuzumab treatment (Figure 6B; Student’s t-test, p = 0.018 and p = 0.014, respectively). Furthermore, the combination of miR-101-5p and lapatinib and trastuzumab caused apoptosis in a significantly greater extent than the miRNA alone (Figure 6B; Student’s t-test, p < 0.001). This is particularly interesting as the targeted drugs alone did not induce apoptosis (Student’s t-test, p = 0.094), indicating that there may be a synergistic effect of the combinatorial treatment.
Discussion
Despite remarkable success of trastuzumab and other targeted therapies, some HER2-positive breast cancers still respond poorly to the treatment offered. This is a challenge, and novel treatment strategies for these patients are highly warranted. We have mimicked these poorly responding breast cancer cases in vitro by using two cell lines that respond poorly to trastuzumab and lapatinib.25 Previously, we have shown that miR-101-5p decrease cell viability in HER2+ breast cancer cells and sensitize the cells to targeted treatment.20 The aim of the current study was to gain knowledge of the mechanisms involved in treatment response, by studying the effects on the proteomic landscape when combining miR-101-5p overexpression with HER2-targeting drugs.
miR-101-5p expression is downregulated in breast cancer,15 non-small cell lung cancer,35 cervical cancer36 and renal cell carcinoma.37 Overexpression of this miRNA inhibited growth in these cancers. This shows that miR-101-5p has tumor-suppressive properties across multiple cancer forms. Our study is, to our knowledge, the first to specifically show the importance of endogenous miR-101-5p expression in HER2-positive breast cancer (Figure 1). Furthermore, we address the proteomic changes upon miR-101-5p overexpression and the effects of this miRNA on apoptosis. Toda et al showed, in HER2-negative cells, the tumor-suppressive properties of miR-101-5p through the targeting of seven putative oncogenic mRNAs,15 none of which are significantly altered or present in our protein data. Thus, our study enlightens other mechanistic properties of miR-101-5p.
We show that miR-101-5p overexpression inhibits cell proliferation in two HER2+ breast cancer cell lines, KPL4 and JIMT1. Further, this miRNA sensitizes poorly responding KPL4 cells to targeted treatment with lapatinib and trastuzumab, leading to decreased cell proliferation and increased apoptosis. It has previously been reported that manipulation of miRNA expression can sensitize cancer cells to targeted therapy, radiation and chemotherapy,38–43 which substantiates the value of using miRNAs to improve the effect of conventional cancer treatment.
We also investigated the effect of combining miR-101-5p with tucatinib, a promising tyrosine kinase inhibitor for HER2 that gained its approval by the Food and Drug Administration for treatment of patients with advanced or metastatic HER2+ breast cancer in 2020. While lapatinib binds both EGFR and HER2, tucatinib has a high selectivity to HER2 compared to EGFR.44 Whereas the addition of trastuzumab and lapatinib significantly improved the effect of miR-101-5p in KPL4 cells, we did not detect the same for tucatinib (Figure 2). Therefore, we speculate that the inhibition of both EGFR and HER2 is important to obtain the additional significant effect of combining the targeted treatments with miR-101-5p. EGFR inhibition has previously shown to decrease viability in KPL4 cells, suggesting EGFR dependency in this cell line.45 Furthermore, lapatinib and trastuzumab did not significantly induce decreased cell viability alone or in combination with miR-101-5p in JIMT1 cells. Thus, the cell line specificity for this combination is evident and further work is needed to fully document the induced biological effects. Nonetheless, miR-101-5p has a significant effect in both cell lines, underlining the potential anti-tumor value of this miRNA.
Alterations in binding domains of HER2, PTEN deficiency and activating mutations in PIK3CA are among the reported mechanisms for resistance to targeted therapy in HER2+ breast cancer.46 PI3K-Akt and mTOR inhibitors, immune checkpoint inhibitors, cell cycle control mechanisms, and blockage of alternative receptor tyrosine kinases such as insulin-like growth factor I (IGF1) receptor have been suggested as potential treatment options for overcoming resistance to targeted therapy.46,47 Another reported resistance mechanism to lapatinib and trastuzumab in HER2+ cells is activation of the estrogen receptor (ER) survival pathway, and treatment can be improved by dual HER2/ER blockage.48 This is, however, not relevant for a significant fraction of HER2+ breast cancer patients, as about 50% are hormone receptor negative.49 KPL4 and JIMT1 cells used in our study represent this patient group, as the cells do not express ER.25
Gale et al have described how targeting ATP can reverse acquired resistance to targeted treatment,50 and interestingly, the level of ATP5A is significantly downregulated in miR-101-5p plus lapatinib and trastuzumab treated KPL4 cells compared to the targeted drugs alone (p = 0.047; Supplementary Table S2). There were no significant changes of this protein level by the mono-treatments. This decrease in ATP5A levels is a potential explanation of how miR-101-5p and the targeted treatment work together to decrease cell proliferation.
HER2 overexpression is associated with suppression of apoptosis,51 and targeted treatment is relevant for overcoming this obstacle. Trastuzumab has been reported to cause apoptosis in HER2-positive breast cancer, in part by reducing the anti-apoptotic protein Mcl-1.52 We do not observe this in trastuzumab resistant KPL4 cells, neither on apoptosis effect (Figure 6B) nor on the levels of Mcl-1 (Supplementary Figure S2, Supplementary Table S2). There is, on the other hand, a significant decrease in Mcl-1 levels by miR-101-5p overexpression alone or in combination with lapatinib and trastuzumab compared to targeted drugs alone (Supplementary Figure S2, Supplementary Table S2). This alteration of Mcl-1 levels contributes to explaining how apoptosis is induced by miR-101-5p and the miRNA plus lapatinib and trastuzumab.
Interestingly, lapatinib was found to induce apoptosis in trastuzumab-resistant SKBR3 HER2+ breast cancer cells by affecting IGF1 signaling.53 This effect is not present in KPL4 cells treated with lapatinib and trastuzumab (Figure 6B), where no significant alterations to the expression of any IGF proteins were observed (Supplementary Table S2). This underlines the complex network of alterations that can be cell type specific and may point to new targets for personalized medicine. The lack of apoptosis upon lapatinib and trastuzumab treatment may in part be due to the intrinsic PTEN deficiency in KPL4 cells25 overriding the apoptosis induction.
Although lapatinib and trastuzumab did not induce apoptosis, the targeted treatment decreased the expression of the anti-apoptotic protein Bcl-xL (Supplementary Figure S2, Supplementary Table S2). Trastuzumab has been reported to potentiate apoptosis by downregulating Bcl-2 expression, but not Bcl-xL in HER2+ breast cancer cells.54 These opposing results exemplify the cellular complexity, where regulation of single proteins is not sufficient for a macroscopic effect on apoptosis. For that reason, miRNA therapeutics may be of particular interest, as one miRNA have multiple mRNA targets. Thus, miRNAs may have a larger impact on the cellular proteomic landscape than targeted treatment, as observed in the protein data presented (Tables 1–3). Although the specific effect of a miRNA may differ between cell lines and cancer types, we here show that miR-101-5p inhibits cell proliferation in two HER2+ breast cancer cell lines. Furthermore, the effects of miR-101-5p on apoptosis have not yet been studied in breast cancer. There are multiple studies on the role of miR-101-3p in apoptosis in multiple cancer forms,10,11,55,56 and one report on miR-101-5p and apoptosis in renal cancer.37 This study is, to our knowledge, the first to report induced apoptosis in breast cancer due to miR-101-5p overexpression.
Conclusion
miR-101-5p expression is downregulated in HER2+ breast cancers. When overexpressed in vitro, miR-101-5p showed tumor-suppressive properties through inhibition of cell proliferation, alteration of protein expression levels relevant for the HER2 signaling pathway, and induction of apoptosis. The miRNA sensitized HER2+ KPL4 breast cancer cells to targeted therapy, suggesting a potential treatment strategy that should be further explored for patients that respond poorly to HER2 therapy.
Acknowledgments
We thank Siri Juell (Oslo University Hospital) for valuable contributions with laboratory experiments and analyses. Further, we want to thank Olav Engebråten (Oslo University Hospital) for his helpful input on the study. The work in this study was in part funded by the South-Eastern Norway Regional Health Authority, project number 2017034 and 2019081. The RPPA Core is supported by NCI Grant # CA16672 and Dr Yiling Lu’s NIH R50 Grant # R50CA221675: Functional Proteomics by Reverse Phase Protein Array in Cancer.
Disclosure
Dr Mads Haugland Haugen reports an international patent application PCT/EP2021/052016 published at WO 2021/156137 A1 https://worldwide.espacenet.com/publicationDetails/originalDocument?CC=WO&NR=2021156137A1&KC=A1&FT=D&ND=3&date=20210812&DB=EPODOC&locale=en_EP. The authors report no other conflicts of interest in this work.
References
1. Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist. 2009;14(4):320–368. doi:10.1634/theoncologist.2008-0230
2. Killelea BK, Chagpar AB, Horowitz NR, Lannin DR. Characteristics and treatment of human epidermal growth factor receptor 2 positive breast cancer: 43,485 cases from the national cancer database treated in 2010 and 2011. Am J Surg. 2017;213(2):426–432.
3. van Maaren MC, de Munck L, Strobbe LJA, et al. Ten-year recurrence rates for breast cancer subtypes in the Netherlands: a large population-based study. Int J Cancer. 2019;144(2):263–272.
4. Baselga J, Cortés J, Kim S-B, et al. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366(2):109–119.
5. Swain SM, Baselga J, Kim S-B, et al. Pertuzumab, trastuzumab, and docetaxel in HER2-positive metastatic breast cancer. N Engl J Med. 2015;372(8):724–734.
6. Murthy RK, Loi S, Okines A, et al. Tucatinib, trastuzumab, and capecitabine for HER2-positive metastatic breast cancer. N Engl J Med. 2019;382(7):597–609.
7. Sun W, Julie Li YS, Huang HD, Shyy JY, Chien S. microRNA: a master regulator of cellular processes for bioengineering systems. Annu Rev Biomed Eng. 2010;12:1–27.
8. Ivey KN, Srivastava D. microRNAs as developmental regulators. Cold Spring Harb Perspect Biol. 2015;7(7):a008144–a008144.
9. O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol. 2018;9:402.
10. Wang L, Li L, Guo R, et al. miR-101 promotes breast cancer cell apoptosis by targeting janus kinase 2. Cell Physiol Biochem. 2014;34(2):413–422. doi:10.1159/000363010
11. Wang R, Wang H-B, Hao CJ, et al. MiR-101 is involved in human breast carcinogenesis by targeting Stathmin 1. PLoS One. 2012;7(10):e46173–e46173. doi:10.1371/journal.pone.0046173
12. Hui Y, Li Y, Jing Y, Feng J-Q, Ding Y. miRNA-101 acts as a tumor suppressor in oral squamous cell carcinoma by targeting CX chemokine receptor 7. Am J Transl Res. 2016;8(11):4902–4911.
13. Li CY, Xiong DD, Huang CQ, et al. Clinical value of miR-101-3p and biological analysis of its prospective targets in breast cancer: a study based on The Cancer Genome Atlas (TCGA) and bioinformatics. Med Sci Monit. 2017;23:1857–1871. doi:10.12659/MSM.900030
14. Liu P, Ye F, Xie X, et al. mir-101-3p is a key regulator of tumor metabolism in triple negative breast cancer targeting AMPK. Oncotarget. 2016;7(23):35188–35198. doi:10.18632/oncotarget.9072
15. Toda H, Seki N, Kurozumi S, et al. RNA-sequence-based microRNA expression signature in breast cancer: tumor-suppressive miR-101-5p regulates molecular pathogenesis. Mol Oncol. 2020;14(2):426–446. doi:10.1002/1878-0261.12602
16. Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. mini review. Front Genet. 2019;10(478). doi:10.3389/fgene.2019.00478
17. Janssen HLA, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi:10.1056/NEJMoa1209026
18. Lindow M, Kauppinen S. Discovering the first microRNA-targeted drug. J Cell Biol. 2012;199(3):407–412.
19. van Zandwijk N, Pavlakis N, Kao SC, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18(10):1386–1396.
20. Normann LS, Aure MR, Leivonen S-K, et al. MicroRNA in combination with HER2-targeting drugs reduces breast cancer cell viability in vitro. Sci Rep. 2021;11(1):10893.
21. Koboldt DC, Fulton RS, McLellan MD, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70.
22. Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352.
23. Goldman M, Craft B, Hastie M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019;326470.doi: 10.1101/326470
24. Kurebayashi J, Otsuki T, Tang CK, et al. Isolation and characterization of a new human breast cancer cell line, KPL-4, expressing the Erb B family receptors and interleukin-6. Br J Cancer. 1999;79(5–6):707–717.
25. Jernstrom S, Hongisto V, Leivonen SK, et al. Drug-screening and genomic analyses of HER2-positive breast cancer cell lines reveal predictors for treatment response. Breast Cancer. 2017;9:185–198.
26. Lu Y, Ling S, Hegde AM, et al. Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol. 2016;43(4):476–483.
27. RStudio Team. RStudio: integrated development for R. Available from: http://www.rstudio.com/.
28. Lingjærde OC, Steen CB, Aure MR, Haakensen VD. Available from https://github.com/cbsteen/clustermap.
29. Krämer A, Green J, Pollard J
30. Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–7.
31. Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14:128.
32. Zou Z, Tao T, Li H, Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell Biosci. 2020;10(1):31.
33. Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, Hynes NE. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc Natl Acad Sci USA. 2003;100(15):8933–8938.
34. Lyu H, Han A, Polsdofer E, Liu S, Liu B. Understanding the biology of HER3 receptor as a therapeutic target in human cancer. Acta Pharm Sin B. 2018;8(4):503–510.
35. Chen Q, Liu D, Hu Z, Luo C, Zheng SL. miRNA-101-5p inhibits the growth and aggressiveness of NSCLC cells through targeting CXCL6. Onco Targets Ther. 2019;12:835–848.
36. Shen W, Xie X, Liu M, Wang L. MicroRNA‐101‐5p inhibits the growth and metastasis of cervical cancer cell by inhibiting CXCL6. Eur Rev Med Pharmacol Sci. 2019;23(5):1957–1968.
37. Yamada Y, Nohata N, Uchida A, et al. Replisome genes regulation by antitumor miR-101-5p in clear cell renal cell carcinoma. Cancer Sci. 2020;111(4):1392–1406.
38. Jiang MJ, Dai JJ, Gu DN, Huang Q, Tian L. MicroRNA-7 inhibits cell proliferation of chronic myeloid leukemia and sensitizes it to imatinib in vitro. Biochem Biophys Res Commun. 2017;494(1–2):372–378.
39. Zawistowski JS, Nakamura K, Parker JS, Granger DA, Golitz BT, Johnson GL. MicroRNA 9-3p targets β1 integrin to sensitize claudin-low breast cancer cells to MEK inhibition. Mol Cell Biol. 2013;33(11):2260–2274.
40. Yan D, Ng WL, Zhang X, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5(7):e11397.
41. Shi L-P, Guo H-L, Su Y-B, Zheng Z-H, Liu J-R, Lai S-H. MicroRNA-149 sensitizes colorectal cancer to radiotherapy by downregulating human epididymis protein 4. Am J Cancer Res. 2018;8(1):30–38.
42. Li LQ, Pan D, Chen Q, et al. Sensitization of gastric cancer cells to 5-FU by microRNA-204 through targeting the TGFBR2-mediated epithelial to mesenchymal transition. Cell Physiol Biochem. 2018;47(4):1533–1545.
43. Eoh KJ, Lee SH, Kim HJ, et al. MicroRNA-630 inhibitor sensitizes chemoresistant ovarian cancer to chemotherapy by enhancing apoptosis. Biochem Biophys Res Commun. 2018;497(2):513–520.
44. Kulukian A, Lee P, Taylor J, et al. Preclinical activity of HER2-selective tyrosine kinase inhibitor tucatinib as a single agent or in combination with trastuzumab or docetaxel in solid tumor models. Mol Cancer Ther. 2020;19(4):976–987.
45. Zhang D, LaFortune TA, Krishnamurthy S, et al. Epidermal growth factor receptor tyrosine kinase inhibitor reverses mesenchymal to epithelial phenotype and inhibits metastasis in inflammatory breast cancer. Clin Cancer Res. 2009;15(21):6639–6648.
46. Pernas S, Tolaney SM. HER2-positive breast cancer: new therapeutic frontiers and overcoming resistance. Ther Adv Med Oncol. 2019;11:1758835919833519.
47. Luque-Cabal M, García-Teijido P, Fernández-Pérez Y, Sánchez-Lorenzo L, Palacio-Vázquez I. Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin Med Insights Oncol. 2016;10(Suppl 1):21–30.
48. Wang Y-C, Morrison G, Gillihan R, et al. Different mechanisms for resistance to trastuzumab versus lapatinib in HER2- positive breast cancers - role of estrogen receptor and HER2 reactivation. journal article. Breast Cancer Res. 2011;13(6):R121.
49. Prat A, Baselga J. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat Clin Pract Oncol. 2008;5(9):531–542.
50. Gale M, Li Y, Cao J, et al. Acquired resistance to HER2-targeted therapies creates vulnerability to ATP synthase inhibition. Cancer Res. 2020;80(3):524–535.
51. Carpenter RL, Lo H-W. Regulation of apoptosis by HER2 in breast cancer. J Carcinog Mutagen. 2013;2013(Suppl 7):3.
52. Henson ES, Hu X, Gibson SB. Herceptin sensitizes ErbB2-overexpressing cells to apoptosis by reducing antiapoptotic Mcl-1 expression. Clin Cancer Res. 2006;12(3 Pt 1):845–853.
53. Nahta R, Yuan LX, Du Y, Esteva FJ. Lapatinib induces apoptosis in trastuzumab-resistant breast cancer cells: effects on insulin-like growth factor I signaling. Mol Cancer Ther. 2007;6(2):667–674.
54. Milella M, Trisciuoglio D, Bruno T, et al. Trastuzumab down-regulates Bcl-2 expression and potentiates apoptosis induction by Bcl-2/Bcl-XL bispecific antisense oligonucleotides in HER-2 gene-amplified breast cancer cells. Clin Cancer Res. 2004;10(22):7747–7756.
55. Guan H, Dai Z, Ma Y, Wang Z, Liu X, Wang X. MicroRNA-101 inhibits cell proliferation and induces apoptosis by targeting EYA1 in breast cancer. Int J Mol Med. 2016;37(6):1643–1651.
56. Wang J, Zeng H, Li H, et al. MicroRNA-101 inhibits growth, proliferation and migration and induces apoptosis of breast cancer cells by targeting sex-determining region Y-box 2. Cell Physiol Biochem. 2017;43(2):717–732.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.