Back to Journals » OncoTargets and Therapy » Volume 8
−148 C/T polymorphism of Axin2 contributes to a decreased risk of cancer: evidence from a meta-analysis
Authors Zhong A, Pan X, Shi M , X H
Received 16 April 2015
Accepted for publication 15 June 2015
Published 29 July 2015 Volume 2015:8 Pages 1957—1966
DOI https://doi.org/10.2147/OTT.S86738
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Daniele Santini
AnYuan Zhong,1 Xue Pan,1 MinHua Shi,1 HuaJun Xu2
1Department of Respiratory Diseases, The Second Affiliated Hospital of Soochow University, Suzhou, 2Department of Otolaryngology, Shanghai Jiao Tong University Affiliated Sixth People’s Hospital, Otolaryngology Institute, Shanghai, People’s Republic of China
Abstract: Several studies have reported an association between −148 C/T polymorphism of Axis inhibition protein 2 (Axin2) and cancer risk; however, the results are inconsistent. In this study, a meta-analysis was performed to assess the association between −148 C/T polymorphism of Axin2 and susceptibility to cancer. Published case–control and cohort-based studies from PubMed, Embase, Wanfang, and CNKI were retrieved, and data were manually extracted. The odds ratios (ORs) and 95% confidence intervals (CIs) of the included studies were pooled. Begg’s and Egger’s tests were used to evaluate publication bias. Cumulative and recursive cumulative meta-analyses (CMA) were performed as evidence accumulated to investigate the trends and stability of the effect size. Nine articles with 1,664 cases and 1,796 controls were included. The pooled effect size showed an association between −148 C/T polymorphism and the risk of cancer (dominant model, OR: 0.72, 95% CI: 0.63–0.83; allele model, OR: 0.81, 95% CI: 0.73–0.90). CMA showed an association trend, and the recursive CMA indicated that more evidence is needed to make conclusions about significance. In a subgroup analysis, a significant association between −148 C/T polymorphism and low cancer susceptibility was detected for lung cancer (dominant model, 0.69, 95% CI: 0.56–0.85; recessive model, OR: 0.75, 95% CI: 0.56–0.99; allele model, 0.76, 95% CI: 0.66–0.86). The −148 C/T polymorphism was also associated with low cancer susceptibility among Asians (dominant model, OR: 0.68, 95% CI: 0.57–0.81; recessive model, OR: 0.75, 95% CI: 0.56–0.99; allele model, OR: 0.76, 95% CI: 0.66–0.86). The Axin2 −148 C/T polymorphism was found to be significantly associated with a decreased risk of cancer, particularly lung cancer, in Asians and population-based controls. Thus, Axin2 should be considered as a potential therapeutic target for preventing tumor growth.
Keywords: cancer, Axin2, gene, polymorphism, meta-analysis
Introduction
As one of the leading causes of death worldwide, cancer is a serious threat to public health1 with approximately 14 million new cases, 8.2 million cancer-related deaths, and 32.6 million people living with cancer (within 5 years of diagnosis) in 2012 (WHO database). Cancer is a multifactorial disease that results from complex gene–gene and gene–environment interactions.2 Genetically, cancer develops as a result of mutational events involving the activation of oncogenes and inactivation of tumor suppressor genes.3 The role of genetic polymorphisms has been widely investigated in many tumor types, and the relationship between tumor suppressor gene polymorphisms and cancer susceptibility has been extensively studied.4 In addition, several previous meta-analyses have evaluated a specific gene polymorphism and risk for all cancer types, respectively.5–7
The Wnt signaling pathway has recently been found to play a central role in human cancer development,8 and Axis inhibition protein 2 (Axin2), a tumor suppressor protein, plays an important role in the Wnt signaling pathway.9 The Axin2 gene contains ten coding exons spanning more than 2.5 kb at human chromosome 17q24 and encodes a protein of 843 amino acids.10,11 Axin2 is a negative regulator of the Wnt signaling pathway; it directs β-catenin for proteasomal degradation.12 A loss of heterozygosity in the genomic locus containing Axin213 and mutations in the gene itself has been observed in certain types of cancer,14–16 suggesting that Axin2 plays an important role in carcinogenesis.
Considering the vital role of Axin2 in carcinogenesis, several case–control studies have been performed to investigate the possible association between -148 C/T (Pro50Ser, rs2240308) polymorphism of Axin2 and cancer susceptibility in different neoplasms and populations.17–25 However, reports on the association between this polymorphism and cancer susceptibility are inconsistent. In addition, the studies published thus far are limited by low statistical power. To shed some light on this controversy, a meticulous meta-analysis was performed to explore the association between -148 C/T polymorphism of Axin2 and cancer risk.
Materials and methods
We performed the meta-analysis according to Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines.26
Publication search
To identify all eligible studies addressing the association between the Axin2 -148 C/T polymorphism and cancer risk, we systematically searched major electronic databases, including PubMed, Embase, Wanfang and CNKI. The NCBI database of genotypes and phenotypes (http://www.ncbi.nlm.nih.gov/gap/) was also searched for Axin2 rs2240308 to identify GWAS data. The last search was updated on May 20, 2015. The following MeSH terms and free words were used: “Axis inhibition protein 2” or “Axin2” compiled with “SNP” or “polymorphism” or “mutation” or “variant” and “tumor” or “cancer” or “carcinoma” or “malignancy”. In addition, we manually searched for references to relevant published studies and review articles. No language restrictions were applied.
Inclusion and exclusion criteria
The inclusion criteria for the meta-analysis were: 1) evaluation of the Axin2 -148 C/T polymorphism and cancer susceptibility; 2) case–control or cohort design; and 3) sufficient data to determine genotype distributions. Studies were excluded for the following reasons: 1) lack of control subjects; 2) non-clinical study; 3) review, abstract, or conference paper; and 4) no reported genotype distribution or allele frequency data.
Data extraction
Two investigators (Drs Zhong and Pan) extracted the required data independently from all eligible studies. The following data were collected from each study: the first author’s name, year of publication, country, ethnicity, genotyping method, P-values from the Hardy–Weinberg equilibrium (HWE) test for the control group, and genotype frequencies among the cases and controls. Discrepancies in data collection were resolved by group discussion. If there were questions or if the details of a particular study were needed, the authors were contacted via e-mail.
Statistical analysis
All statistical analyses were performed using Stata (ver. 11.0; StataCorp LP, College Station, TX, USA). Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the association between the Axin2 -148 C/T polymorphism and cancer risk in a dominant model (TT + CT vs CC), recessive model (TT vs CT + CC), and allele model (T vs C allele). In cases in which an included study made adjustments for various covariates, the most fully adjusted OR was used. Subgroup analyses were performed according to cancer type, ethnicity, and source of control. Homogeneity was tested using the Q-statistic and I2 statistic if P>0.10. The Mantel-Haenszel fixed-effects model was used when no heterogeneity existed among the studies; otherwise, DerSimonian and Laird’s random-effects model was used.27,28 HWE of the genetic frequency distributions for the controls was calculated using Pearson’s chi-square test. The significance of the pooled OR was determined by a Z-test. Begg’s and Egger’s test were used to evaluate publication bias.29,30 A sensitivity analysis was performed by the leave-one-out procedure.31 Unless otherwise stated, a P-value <0.05 was considered statistically significant. Cumulative meta-analyses (CMA) and recursive CMA were performed for each polymorphism to evaluate the trend of the risk effect (ie, OR) of the allele contrast over time.32 For the CMA, studies were chronologically ordered by publication year, and the risk effect was obtained at the end of each year/information step. In the recursive CMA, the relative change in pooled OR at each information step was calculated.
Results
Search results
After searching the aforementioned electronic databases, a total of 815 references were obtained. Duplicate studies were excluded, leaving 690 papers. Among these, 598 papers were excluded because they did not meet our inclusion criteria. Another 83 articles were excluded because they lacked controls (n=24), had insufficient data (n=53), or did not include -148 C/T polymorphism of Axin2 (n=6). Ultimately, nine articles were included in our meta-analysis and subjected to further statistical analysis (Figure 1).
  | Figure 1 Flow chart of the literature search and study selection process. |
Study characteristics
Table 1 summarizes the characteristics of the included studies. Publication year ranged from 2006 to 2014. All nine articles, including eleven studies (1,664 patients and 1,796 controls), examined -148 C/T Axin2 polymorphism and cancer risk.17–25 Of the nine articles, four were from Asia,18,19,23 three were from the People’s Republic of China,19,23,24 and one was from Japan.20 The others were from western countries, one from Poland,23 and three from Turkey.17,21,22 Regarding the ethnicity of the subjects, six studies included subjects of Asian origin18,19,23,25 and five included Caucasian subjects.17,20–22,25 Of the eleven included studies, ten used the polymerase chain reaction-restriction fragment length polymorphism technique17,18,20–25 and one used the TaqMan method.19 Of the eleven studies, two considered prostate cancer,21,22 three considered lung cancer,17–19 two considered colorectal cancer,23,24 one considered head and neck cancer,23 one considered ovarian cancer,20 one considered astrocytoma,22 and one considered papillary thyroid cancer cancer.25 All the controls in the included studies were in HWE.
Quantitative synthesis
In dominant, recessive, and allele models, no significant between-study heterogeneity was found among the included studies (I2=0.0%, P=0.62; I2=9.2%, P=0.356; and I2=5.2%, P=0.394, respectively), so a fixed-effects model was used to analyze the data. The pooled data indicated a significant association between -148 C/T polymorphism of Axin2 and a decreased risk of cancer using a dominant model and an allele model (OR: 0.72, 95% CI: 0.63–0.83; OR: 0.81, 95% CI: 0.73–0.90, respectively) (Figures 2 and 3). The Axin -148 C/T polymorphism was not significantly associated with cancer risk in the recessive model (OR=0.84, 95% CI 0.69–1.02) (Figure 4).
Subgroup analyses
Subgroup analyses were performed to investigate the effects of cancer type, ethnicity, and source of control. In the cancer type-specific subgroup analysis, the Axin2 -148 C/T polymorphism was associated with a decreased risk of lung cancer (dominant model, OR: 0.69, 95% CI: 0.56–0.85; recessive model, OR: 0.61, 95% CI: 0.44–0.85; and allele model, OR: 0.73, 95% CI: 0.63–0.85). When stratified by ethnicity, a significant association between the Axin2 -148 C/T polymorphism and decreased cancer risk was identified in Asians dominant model, OR: 0.68, 95% CI: 0.57–0.81; recessive model, OR: 0.75, 95% CI: 0.56–0.99; and allele model, OR: 0.76, 95% CI: 0.66–0.86. Similarly, this association was found in the population-based (PB) controls (dominant model, OR: 0.72, 95% CI: 0.60–0.88; recessive model, OR: 0.72, 95% CI: 0.53–0.98; and allele model, OR: 0.78, 95% CI: 0.68–0.90). The main results of the pooled estimates in this meta-analysis are presented in Table 2. In addition, we also performed a subgroup analysis in lung cancer patients (Table 3).
  | Table 2 Subgroup analysis of the studies included in this meta-analysis |
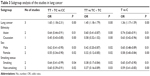  | Table 3 Subgroup analysis of the studies in lung cancer |
Sensitivity analysis
To evaluate the influence of each included study, a sensitivity analysis was performed. The leave-one-out procedure was used to estimate the contribution of each study to the pooled OR value.33 For all cancer types, the ORs ranged from 0.70 (95% CI: 0.61–0.81) to 0.82 (95% CI: 0.74–0.91). For lung cancer, the ORs ranged from 0.39 (95% CI: 0.21–0.73) to 0.76 (95% CI: 0.64–0.90). The statistical significance of the results was not altered when any single study was omitted. Therefore, the stability of the study was not influenced by any individual study.
Publication bias
Begg’s and Egger’s tests were used to evaluate publication bias. No significant evidence of publication bias was found in the dominant, recessive, or allele models (Begg’s test: P=0.88, 0.76, and 1.00, respectively; Egger’s test: P=0.60, 0.81, and 0.76, respectively). The shapes of the funnel plots for the association between -148 C/T polymorphism and cancer risk also revealed no evidence of obvious asymmetry in the dominant, recessive, or allele models (Figures 5–7). In the CMA, the pooled genetic risk effect remained significant during the entire study period. However, instability in pooled OR change was seen in the recursive CMA (Figure 8). Thus, an association trend exists, and more evidence is needed to draw a safe conclusion on the significance and magnitude of the effect size.
  | Figure 5 Funnel plots of Axin2 -148 C/T polymorphism in the dominant model. |
  | Figure 6 Funnel plots of Axin2 -148 C/T polymorphism in the recessive model. |
  | Figure 7 Funnel plots of Axin2 -148 C/T polymorphism in the allele model. |
Discussion
In the present meta-analysis, we investigated the association between -148 C/T polymorphism of Axin2 and cancer risk. Our results indicate an association between -148 C/T polymorphism and a decreased risk of cancer in the dominant, recessive, and allele models. A subgroup analysis revealed that -148 C/T polymorphism was associated with a decreased risk of lung cancer in all models and prostate cancer in the dominant model, while colorectal and other cancers were unrelated to -148 C/T polymorphism of Axin2. Ethnicity-based investigations revealed that the dominant, recessive, and allele models of -148 C/T in Asians contributed to a decreased risk of cancer. Moreover, when data were stratified by the source of control, there was an association between subgroup and PB controls in all models. A significant association was found in all lung cancer subgroups, and data were grouped by ethnicity (Caucasian and Asian), sex, and smoking status.
It was previously reported that the Axin2 -148 C/T genotype is associated with a decreased risk of cancer. One study suggested that this polymorphism could be used as a marker to identify individuals with a low risk of developing lung cancer.18 A recent study reported an association between -148 C/T genotype and a low risk of prostate cancer.23 Based on our meta-analysis, we speculate that -148 C/T polymorphism of Axin2 alters transcription of the gene, thereby decreasing the risk of the onset of cancer. However, such an association has not been reported in colorectal cancer,18,24 head and neck cancer,18 ovarian cancer,20 astrocytoma,2,22 or papillary thyroid cancer.25 This may be partly explained by the limited number of studies conducted and the small sample size included in each study.
The Axin2 gene is known to have a single nucleotide polymorphism (SNP) at codon 50 encoding either proline (CCT) or serine (TCT). To date, the biological mechanism linking Axin2 to carcinogenesis has not been clarified. Axin2 is a multi-domain scaffold protein that plays an important role in various biological processes, including Wnt/β-catenin signaling.33,34 Aberrant Wnt/β-catenin signaling occurs in various cancers.35 A large complex of Axin, adenomatous polyposis coli, β-catenin, GSK3β, and several other proteins regulates Wnt signaling through the regulation of β-catenin cytoplasmic localization.16,36 The predicted functional domains of the Axin2 protein include the regulator of G protein signaling (RGS) domain (amino acids 81–200), the GSK3 interaction domain (amino acids 327–413), and the β-catenin binding site (amino acids 413–476).18 The RGS domain has been reported to function as the adenomatous polyposis coli-binding site. The RGS domain (amino acids 81–200) participates in Axin2 tumor suppressor functions.37 The location of the Axin2 rs2240308 SNP at codon 50 is extremely close to the RGS domain.18 Thus, genetic variation in Axin2 may affect the risk of developing cancer.
During the review process of this manuscript, a meta-analysis evaluating rs2240308 and the risk of cancer was published;38 however, the meta-analyses in the present study provide much more information. The differences between this recently published report and our study are as follows: 1) we included more studies that met our inclusion and exclusion criteria; thus, the results might be more accurate; 2) the previous meta-analysis only performed a subgroup analysis based on ethnicity and cancer type; we performed not only a subgroup analysis stratified by ethnicity and cancer type but also lung cancer-specific subgroup analyses by ethnicity, sex, and smoking status in PB or hospital-based controls; and 3) we performed additional CMA and recursive CMA to investigate the trend and stability of effect sizes as evidence accumulated. The CMA for Axin2 showed a consistent and significant trend toward an association as evidence from published studies accumulated. The recursive CMA indicated instability in the relative change in risk effects, and therefore, more evidence is needed to claim or deny the existence of an association between the variants and cancer.
Some limitations of this meta-analysis should be addressed. First, the meta-analysis was based on unadjusted risk estimates. A lack of data on possible effect modifiers (eg, sex, age, and clinical manifestations) in the included studies may have affected the identification of associations with disease. Second, sampling variability within each study may be a confounding factor in determining the effect of genetic variation on cancer susceptibility. Third, the sample size was relatively small, restricting the power and subgroup analysis of this meta-analysis. Fourth, the etiology of common disorders is multifactorial and involves complex epistemic and gene–environment interactions. Nevertheless, this is the first meta-analysis to explore the relationship between -148 C/T polymorphism of Axin2 and cancer risk. The major findings of this meta-analysis were a lack of heterogeneity and no publication bias.
Conclusion
In summary, the present meta-analysis provides clear evidence showing that -148 C/T polymorphism of Axin2 contributes to a decreased risk of cancer, especially lung cancer, in Asians and PB controls. However, studies of gene–gene and gene–environment interactions should be considered in the future to obtain a more comprehensive understanding of the association between the Axin2 -148 C/T polymorphism and cancer risk. A future study that consists of a larger sample size is required to further evaluate this association.
Acknowledgment
Our studies were supported by a grant from the National Natural Science Foundation of China (no 81272610).
Author contributions
Both AnYuan Zhong and Xue Pan contribute equally to this paper. This paper is approved by all authors for publication. All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors have no conflicts of interest to declare.
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics. CA Cancer J Clin. 2014;64(2):9–29. | ||
Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–860. | ||
Chiao PJ, Bischoff FZ, Strong LC, Tainsky MA. The current state of oncogenes and cancer: experimental approaches for analyzing oncogenetic events in human cancer. Cancer Metastasis Rev. 1990;19(1):63–80. | ||
Miremadi A, Oestergaard MZ, Pharoah PDP, Caldas C. Cancer genetics of epigenetic genes. Hum Mol Genet. 2007;16:28–49. | ||
Zhang X, Weng WH, Xu W, et al. The association between the migration inhibitory factor −173g/c polymorphism and cancer risk: a meta-analysis. Onco Targets Ther. 2015;8:601–613. | ||
Cheng DY, Hao YW, Zhou WL. IL-1α −889 c/T polymorphism and cancer susceptibility: a meta-analysis. Onco Targets Ther. 2014;7:2067–2074. | ||
Dai W, Zhou Q, Tan XX, Sun CF. IL-17A (−197G/A) and IL-17F (7488T/C) gene polymorphisms and cancer risk in Asian population: a meta-analysis. Onco Targets Ther. 2014;7:703–711. | ||
Neth P, Ries C, Karow M, Egea V, Ilmer M, Jochum M. The Wnt signal transduction pathway in stem cells and cancer cells: influence on cellular invasion. Stem Cell Rev. 2007;3(1):18–29. | ||
Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15(1):28–32. | ||
Mai M, Qian C, Yokomizo A, Smith DI, Liu W. Cloning of the human homolog of conductin (AXIN2), a gene mapping to chromosome 17q23-q24. Genomics. 1999;55(3):341–344. | ||
Salahshor S, Woodgett JR. The links between Axin and carcinogenesis. J Clin Pathol. 2005;58(3):225–236. | ||
Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22(4):1172–1183. | ||
Phelan CM, Borg A, Cuny M, et al. Consortium study on 1280 breast carcinomas: allelic loss on chromosome 17 targets subregions associated with family history and clinical parameters. Cancer Res. 1998;58(5):1004–1012. | ||
Taniguchi K, Roberts LR, Aderca IN. Mutational spectrum of beta-catenin, Axin1 and Axin2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21(31):4863–4871. | ||
Wu R, Zhai Y, Fearon ER, Cho KR. Diverse mechanisms of beta-catenin deregulation in ovarian endometrioid adenocarcinomas. Cancer Res. 2001;61(22):8247–8255. | ||
Liu W, Dong X, Mai M, et al. Mutations in Axin2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TcF signaling. Nat Genet. 2000;26(2):146–147. | ||
Gunes EG, Pinarbasi E, Pinarbasi H, Silig Y. Strong association between lung cancer and the AXIN2 polymorphism. Mol Med Rep. 2009;2(6):1029–1035. | ||
Kanzaki H, Ouchida M, Hanafusa H, et al. Single nucleotide polymorphism of the AXIN2 gene is preferentially associated with human lung cancer risk in a Japanese population. Int J Mol Med. 2006;18(2):279–284. | ||
Liu D, Li L, Yang Y, Liu W, Wu J. The Axin2 rs2240308 polymorphism and susceptibility to lung cancer in a Chinese population. Tumor Biol. 2014;35(11):10987–10991. | ||
Mostowska A, Pawlik P, Sajdak S, et al. An analysis of polymorphisms within the Wnt signaling pathway in relation to ovarian cancer risk in a Polish population. Mol Diagn Ther. 2014;18(1):85–91. | ||
Pinarbasi E, Gunes EG, Pinarbasi H, Donmez G, Silig Y. AXIN2 polymorphism and its association with prostate cancer in a Turkish population. Med Oncol. 2011;28(4):1373–1378. | ||
Gunes EG, Pinarbasi E, Pinarbasi H. AXIN2 polymorphism and its association with astrocytoma in a Turkish population. Mol Med Rep. 2010;3(4):705–709. | ||
Ma C, Liu C, Huang P, et al. Significant association between the Axin2 rs2240308 single nucleotide polymorphism and the incidence of prostate cancer. Oncol Lett. 2014;8(2):789–794. | ||
Naghibalhossaini F, Zamani M, Mokarram P, Khalili I, Rasti M, Mostafavi-pour Z. Epigenetic and genetic analysis of WNT signaling pathway in sporadic colorectal cancer patients from Iran. Mol Biol Rep. 2012;39:6171–6178. | ||
Liu X, Li S, Lin XJ, Yan KK, Zhao LY, Bao HH. Association between ITGA3, TP53INP2, and AXIN2 gene polymorphism and papillary thyroid carcinoma. J Jilin Univ. 2014;40:384–388. | ||
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006–1012. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Ylmaz M, Bayazit YA, Ciftci TU, et al. Association of serotonin transporter gene polymorphism with obstructive sleep apnea syndrome. Laryngoscope. 2005;115(5):832–836. | ||
Zintzaras E, Lau J. Synthesis of genetic association studies for pertinent gene–disease associations requires appropriate methodological and statistical approaches. J Clin Epidemiol. 2008;61:634–645. | ||
Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17(5):1371–1384. | ||
Furuhashi M, Yagi K, Yamamoto H, et al. Axin facilitates Smad3 activation in the transforming growth factor beta signaling pathway. Mol Cell Biol. 2001;21(15):5132–5141. | ||
Herr P, Hausmann G, Basler K. Wnt secretion and signalling in human disease. Trends Mol Med. 2012;18(8):483–493. | ||
Behrens J, Jerchow BA, Würtele M, et al. Functional interaction of an Axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280(5363):596–599. | ||
Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. | ||
Gong J, Jiang Y, Hao NB, Zhu B, Li YS. Quantitative assessment of the association between AXIN2 rs2240308 polymorphism and cancer risk. Sci Rep. 2015;5:10111. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





