Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNAs: Multifaceted Regulators of Colorectal Cancer Metastasis and Clinical Applications
Authors Wen XQ , Qian XL , Sun HK, Zheng LL , Zhu WQ, Li TY, Hu JP
Received 1 June 2020
Accepted for publication 12 September 2020
Published 27 October 2020 Volume 2020:13 Pages 10851—10866
DOI https://doi.org/10.2147/OTT.S265580
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Tohru Yamada
Xiang-Qiong Wen,1,* Xian-Ling Qian,2,3,* Huan-Kui Sun,1 Lin-Lin Zheng,1 Wei-Quan Zhu,1 Tai-Yuan Li,1 Jia-Ping Hu1
1Department of General Surgery, The First Affiliated Hospital of Nanchang University; Medical College of Nanchang University, Nanchang, Jiangxi, 330006, People’s Republic of China; 2Department of Radiology, Zhongshan Hospital, Fudan University, Shanghai, 200032, People’s Republic of China; 3Department of Medical Imaging, Shanghai Medical College,Fudan University, Shanghai, 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jia-Ping Hu Email [email protected]
Abstract: Colorectal cancer (CRC) is the third-commonest malignant cancer, and its metastasis is the major reason for cancer-related death. The process of metastasis is highly coordinated and involves a complex cascade of multiple steps. In recent years, miRNAs, as highly conserved, endogenous, noncoding, single-stranded RNA, has been confirmed to be involved in the development of various cancers. Considering that miRNA is also involved in a series of biological behaviors, regulating CRC occurrence and development, we review and summarize the role of miRNAs and related signaling pathways in several CRC-metastasis stages, including invasion and migration, mobility, metabolism, epithelial–mesenchymal transition, tumor-microenvironment communication, angiogenesis, anoikis, premetastatic–niche formation, and cancer stemness. In addition, we review the application of miRNAs as diagnostic CRC markers and in clinical treatment resistance. This review can contribute to understanding of the mechanism of miRNAs in CRC progression and provide a theoretical basis for clinical CRC treatment.
Keywords: colorectal cancer, miRNA, metastasis, invasion, migration, stemness
Introduction
Colorectal cancer (CRC) is the third-commonest of all cancers, with an estimated 145,600 new cases worldwide in 2019. Next only to lung cancer and breast cancer, CRC accounts for 8%–9% of all cancers. In terms of its mortality rate, it ranks third in males and second in females.1 TNM stage is the main indicator of CRC prognosis, and early tumor stage indicates better survival. Unfortunately, due to lack of early symptoms, a considerable proportion of patients are diagnosed at an advanced stage and even coexist with metastasis. Comprehensive CRC-treatment therapy, including surgery, radiotherapy, chemotherapy and immunity therapy and neoadjuvant chemotherapy, has improved the 5-year survival rate significantly,2 though many CRC patients still experience high risks of tumor recurrence and metastasis. Approximately 60% of patients with distant metastasis and even 20% patients were found the formation of multiple colorizations after comprehensive treatment in 5 years.3 The liver and peritoneum are the commonest sites of distant CRC metastasis.4 Therefore, understanding the mechanism of CRC metastasis can be of great importance.
The CRC-metastasis cascade is a set of complex and highly coordinated processes. First, CRC cells undergo a series of continuous evolution processes that endow more capacity for invasion and migration, and CRC cells are inclined to adopt an aggressive phenotype. These reprogramming processes including mobility alteration, metabolism regulation, and epithelial–mesenchymal transition (EMT) of CRC cells.5 Second, CRC cells destroy basement membrane and extracellular matrix (ECM), then interact with noncancer cells in the tumor microenvironment (TME).6 Third, the angiogenesis and lymphangiogenesis are induced, which work as an aisle for distant metastasis. CRC cells also promote vascular permeability boost cell leakage into the bloodstream and penetration in distant tissue.7–9 Above processes promote the generation of circulating tumor cells (CTCs). Fourth, the CTCs acquire anoikis resistance and survive the immune system during circulation. This acts as a precondition for distant primary cancer–cell metastasis. Then, the primary cancer cells promote the formation of a premetastatic niche (PMN) to facilitate the adhesion of CTCs to vascular endothelium and extravasation to distant organs and tissue. Finally, CRC cells reach metastatic sites and generate second colonization.10 The metastasis processes are not sequential. Instead, they intersect in time and spatial dimension. For example, primary tumor tissue continues to release CTCs into the circulation, which can even be detected in very early on in the TNM process, and vessel structure is also destroyed during the ECM-degradation process.11,12 Cancer stem cells (CSCs) are viewed as the instigators of growth, resistance to chemotherapy, and recurrence and metastasis of cancer. The stemness of CSCs is the major challenge in the fight against CRC recurrence and metastasis, and the elimination of CSCs is believed to be a potential method to overcome malignant disease.13
miRNAs are members of the endogenous noncoding RNA (ncRNA) family, with a length of approximately 22 nucleotides.14 The generation of miRNAs involves pri-miRNA catalysis by RNA polymerase II in the nucleus and a series of modifications by RNase III endonuclease and Dicer.15 They bind to the 3′-untranslated regions (3′-UTRs) of target protein-encoding mRNAs and form RNA–RNA complexes, resulting in translation suppression or mRNA degradation. One miRNA can regulate multiple target genes, and one gene can be regulated by many miRNAs synchronously. It is estimated that about 60% of protein-coding genes are controlled by various miRNAs.16 Accumulating evidence has revealed that dysregulation of miRNAs is associated with multiple biological processes, such as proliferation, differentiation, apoptosis, and autophagy.17 As for cancer, miRNAs participate in initiation, progression, and metastasis. In the CRC-metastasis cascade, miRNAs act as oncogenes or tumor-suppressive factors; therefore, here we summarize state-of-the-art advances and explore the underlying miRNA-related molecular mechanisms of CRC metastasis. In addition, we review the application of miRNAs as diagnostic CRC markers and in drug resistance, hoping to provide a theoretical basis for clinical CRC treatment.
miRNAs, Invasion, and Migration
miRNAs and Mobility
CRC cells undergo a series of distinct reprogramming processes to increase mobility, thus promoting cell invasion and migration.18 During these processes, CRC cells speed up cytoskeleton reconstruction and change cell morphology, which facilitate cell crawling, deforming, squeezing, through narrow tissue gaps, and crossing vessel walls.19 Connexins exist on cell surfaces and interact with the surrounding environment. After reprogramming processes, the adhesion pattern is altered and cells are able to detach from the primary tumor niche.20 During cell movement, a polar foot protrusion termed lamellipodia or filopodia will be formed at the cell’s leading edge.21 In an anoxic environment, through activating the RECK–FAK–Akt–Rac1 signaling pathway, the hypoxia-sensitive miR590-5p enhances CRC-cell sprouting and accelerates the formation of protrusions, and miR590-5p inhibitors can impact cell mobility conversely.22
A remarkable rearrangement of cytoskeleton facilitates cell motility. The RhoA–ROCK signaling network is a critical signaling pathway in cytoskeleton regulation.19,23 miR133a-3p acts as an inhibitor, and its expression is reduced in CRC. Low expression of miR133a-3p in CRC cells activates the RhoA–ROCK signaling network, subsequently promoting cytoskeleton remodeling.24 By inhibiting SLAIN2, miR106b-5p impairs mobility via blocking microtubule dynamics. Low miR106b-5p levels have been demonstrated to promote the mobility of CRC cells.25 DLC1 is recognized as a member of Rho GTPase–activating protein family. CRC cell–derived exosomal miR106b-3p promotes cytoskeleton rearrangement by suppressing DLC1.26
As a critical downstream-signaling molecule of the RhoA–ROCK pathway, cofilin enhances actin-filament remodeling and promotes mobility.19 PAK4 is a serine/threonine kinase that regulates the LIMK1–cofilin pathway. miR145 impairs mobility of CRC cells by suppressing PAK4 and inactivating this pathway.27 miR145 can also impair CRC-cell mobility via inhibiting the expression of MYO6.28 CRC cells change adhesion pattern by regulating connexin expression. Integrins are major molecules that mediate the connection between CRC cells and the ECM.29,30 In addition to being involved in EMT and proliferation, miR27b-3p in CRC cells promotes an aggressive morphology and intercellular junction pattern by suppressing HOXA10, thus increasing integrin β1 expression. It also results in cytoskeleton remodeling and a spindle-shaped phenotype.31 Similarly, miR130b has been found to bind directly to the 3ʹ-UTR of integrin β1 mRNA.32 miR30e-5p suppresses metastasis by directly binding to mRNAs of integrin α6 and integrin β1. However, controlled by p53, miR30e-5p is at a significantly low level in CRC cells.33 The tumor-suppressive miR143-3p has also been found to directly inhibit the expression of integrin α6. miR330-5p and miR19b-3p inhibit integrin α5 and integrin β8, respectively, to promote cancer metastasis.34,35
miRNAs and Metabolism
Cancer cells undergo a form of metabolic reprogramming termed the Warburg effect. Even under conditions of sufficient oxygen supply, cancer cells incline to ingest a lot of glucose and undergo anaerobic sugar metabolism, produce lactic acid. This process sustains energy supply for cell migration, and the production of lactic acid creates a prometastasis environment.36 Overexpression of miR27a relates to the silence of mitochondrial activity and exhibits great glycolysis potential. miR27a in CRC cells significantly enhances metabolic reprogramming via interacting with metabolism-related genes, including hexokinase, GCK, pyruvate dehydrogenase, MAPK, PTEN, and PI3K.37 DKK2 is a kind of secreted protein and is highly expressed in metastatic cancer. DKK2 can accelerate aerobic glycolysis speed, and low levels of miR493-5p in CRC cells promote the production of lactic acid via activating the DKK2–PI3K–Akt–mTOR pathway. Reexpression of miR493-5p has been found to repress glucose uptake and reduce the production of lactic acid.38 Expression of PKM2 is related to the dysregulation of miR122. By inhibiting the expression of PKM2, miR122 affects the glycolysis rate. In CRC, the expression of miR122 is repressed and results in an overexpression of PKM2, which induces anaerobic sugar metabolism.39
miRNAs and EMT
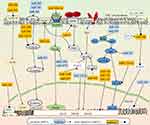
EMT is a vital process in the cancer-metastasis cascade that endows cancer cells with great potential for metastasis. During EMT, cancer cells lose the epithelial phenotype and acquire the mesenchymal phenotype. In detail, E-cadherin in cancer cells is downregulated, thus losing apical–basal polarity and the original tight connection of cells, whereas mesenchymal-associated markers are upregulated, mainly N-cadherin, vimentin, and fibronectin, and then obtain an invasive cell morphology.40 EMT is triggered or silenced in response to diversity stimulation and intracellular components.5 In the TME, the expression of miRNAs is diverse. Through controlling the expression of the EMT-related core transcription factors, such as Snail, Slug, ZEB, and several signaling pathways, miRNAs orchestrate complex biological events.41 We summarize the miRNA-related EMT-regulating network in CRC cells in Figure 1 and give some new examples.
HIF1α and TP53 conversely control the EMT/MET process through miR34a. Under hypoxic conditions, HIF1α in TP53-defective CRC cells directly represses the expression of miR34a, which promotes hypoxia-induced EMT via activating STAT3. In contrast, under hypoxic conditions but with the presence of sufficient TP53 expression, expression of miR34a is enhanced, and overexpression of miR34a leads to the suppression of hypoxia-induced EMT.42 The suppression of miR34a and activation of the STAT3 pathway shift the cell phenotype toward a mesenchymal state and enhance the metastatic cascade. On the contrary, highly expressed miR34a and an inactivated STAT3 pathway switch back the cell phenotype to an epithelial state and allow colonization of metastases.
Endogenous miRNAs exist mainly in cytoplasm. They also present extracellularly via protein carriers, liposome carriers, and exosomes. Exosomes are small vesicles of 30–100 nm in size and often work as transport vesicles between various cells. They are enriched with signal molecules, including proteins, lipids, DNA, RNA, and miRNAs.43 When they are released by cells and taken up by others, they participate in multiple biological events.44 DLC1 is a member of the Rho GTPase–activated protein family and controls the expression of EMT-associated proteins.45 Highly metastatic CRC cells release miR106b-3p–contained exosomes to less metastatic CRC cells. The oncogenic miR106b-3p in the latter increases N-cadherin and decreases E-cadherin at the protein level via suppressing DLC1. Collectively, oncogenic miR106b-3p–containing exosomes promote the invasive capacity of low invasive–potential cells and endow CRC cells in situ more capacity for metastasis. High levels of miR106b-3p exosomes in serum indicate poor prognosis, and miR106b-3p inhibitors impair cell-migration capacity.26
miR126-5p works as a tumor-suppressive miRNA and is lowly expressed in CRC. It interacts with the 3′-UTRs of Slug, Twist, and VEGFα mRNAs, leading to inhibition of the EMT process and angiogenesis in CRC. Further exploration has demonstrated that the lncRNA, MALAT1 binds directly to miR126-5p and impairs its function. The potent oncogenic factor YAP1 promotes the EMT process and angiogenesis by inhibiting the tumor-suppressive miR126-5p.46 A bulk of miRNAs are involved in the regulation of EMT. They work as oncogenes or tumor-suppressive regulators in tumor progression by controlling downstream targets. miR34a and miR598 suppress EMT by inhibiting the Notch pathway. miR598 and miR10b play a negative role by regulating cMyc, K-Ras, and KLF4. miR582, miR183, miR496, and miR452 positively enhance EMT by directly or indirectly activating the Wnt–β-catenin pathway, while miR200c, miR206, and miR139-5p suppress EMT by inhibiting GSK3β. Although miR30a, miR9, miR1249, miR497, and miR488 act on different proteins or receptors, they inhibit EMT by MAPK. Overexpression of miR4775 and miR1269 promotes EMT and induces an aggressive phenotype via activating the TGFβ–Smads pathway, while miR500a-5p plays a negative role via this signaling pathway. miR105 and miR183 promote EMT by activating the PI3K–Akt pathway, while miR1224-5p inhibits EMT via this pathway. miR34a plays a negative role in EMTvia inhibiting the STAT3 pathway. To sum up, these signaling pathways, molecules, and some miRNAs, such as miR138, miR215, and miR145 target EMT-related core transcription factors and regulate EMT in CRC. In Table 1, some tumor-suppressive and oncogenic miRNAs and their direct targets are summarized. Those miRNAs promote or inhibit EMT by positively or negatively controlling downstream-signaling cascades. Relationships between miRNAs and signal cascades are depicted in Figure 1.
 |
Table 1 Dual effects of miRNAs in CRC EMT process |
miRNAs and TME
The TME comprises the ECM, immune cells, fibroblasts, various signaling molecules, and blood vessels. The ECM consisted mainly of collagen, elastin, and proteoglycans and usually acts as a barrier to cancer metastasis.6 There are numerous types of non-TCs in the TME, including cancer-associated fibroblasts (CAFs), endothelial cells, and inflammatory cells. Cross talk between cancer cells and non-TCs participates in metastasis.6,76,77 CRC cells destroy the ECM and make passageways via secreting such substances as proteolytic enzymes in the form of MMP. For example, miR194 in CRC cells promotes metastasis via inducing the expression of MMP2.54 High miR203 level scan attenuate the metastatic potential of CRC by blocking the ERK–MMP9 pathway.78 miR139-5p impairs metastasis of CRC via inhibiting the Notch pathway and suppressing the expression of MMP7 and MMP9.79 ADAMTS5 shares the domains of MMP and possesses proteolytic enzyme–like functions. Due to downregulation of miR140-5p, the of ADAMTS5 expression is increased, leading to ECM destruction.80
Studies have shown that CAFs can promote CRC metastasis by interacting with CRC cells through miRNAs. For example, miR92a-3p–contained exosomes are released by CAFs and taken up by CRC cells, thus activating the intracellular Wnt–β-catenin pathway by inhibiting FBXW7 and MOAP1.81 Also, CAFs can attenuate the inhibitor role of miR141 by releasing exosomal lncRNA H19. The lncRNA H19 from CAFs can activate the Wnt–β-catenin pathway and promote stemness by sponging miR141 in CRC cells.82 Tumor-associated macrophages (TAMs) are the most abundant immune cells in the TME, and M2 phenotype TAMs promote cancer progression and take part in ECM remodeling and immuno response. On the one hand, TAMs can promote invasion and migration of CRC cells. TAMs inhibit the expression of miR506-3p in CRC, and in turn accelerate EMT process by activating FoxQ1. Meanwhile, mesenchymal phenotype CRC cells enhance the recruitment of TAMs via secreting CCL12.77 On the other hand, TAMs can also be stimulated by CRC cells. The tumor suppressor miR195-5p is decreased in CRC cells, and low miR195-5p levels induce EMT by activating the Notch2 pathway. Meanwhile, dysregulation of miR195-5p enhances the reprogramming of TAMs by stimulating CRC cells to secrete IL4.76
miRNAs and Angiogenesis
With uncontrolled cell division and growth of cancer tissue, a hypoxic environment is formed inside the tumor. The key HIF1 is upregulated in response to hypoxia, which boosts the expression of angiogenesis-related genes, including ANG2, HIF1, TGFB, VEGF, and PLGF.83 miRNAs control angiogenesis at multiple levels via suppressing antiangiogenesis genes or promoting proangiogenesis genes.7 miRNAs can affect the response to hypoxia of CRC through regulating the expression of HIF1. Induced by chemokine CCL19, miR206 inhibits the Met–ERK–Elk1 pathway and suppresses the expression of HIF1α, leading to the inhibition of angiogenesis in CRC.84 Similarly, miR107 regulates hypoxia signaling by inhibiting the expression of HIF1β. Downregulation of miR107 has been found to accelerate neovascularization.85 miRNAs mediate the regulation of VEGF expression and play a critical role in promoting CRC angiogenesis.86 CRC cell–derived miR1229 accelerates angiogenesis through activating the HIPK2–VEGFα–VEGFR pathway, and high levels of exosomal miR1229 in serum have been demonstrated to enhance tube formation in vitro and accelerate angiogenesis in vivo.87
CRC-derived miRNA-contained exosomes can facilitate metastasis by increasing vascular permeability. KLF4 is an oncogenic transcription factor of cell junction related protein which supports the integrity of endothelial barriers.88 miR25-3p from CRC cells targets endothelial cells and significantly decreases the expression of cell-cell junction related protein (ZO-1, occludin, Cldn5) via suppression of KLF4. This results in a significant increase of CRC cells leakage to bloodstreams and facilitates distant metastasis. In further, miR25-3p makes a similar changing in the formation of PMN and accelerates extravasation.8 Analogously, miR92a from CRC cells decreases cell-cell tight connection components and cell adhesion. And it also stimulates cell mitosis and accelerates micro-tube formation to promote angiogenesis.89 Lymphatic metastasis is also a common way of metastasis, therefore, lymphangiogenesis is also critical.9 VEGFC/D/VEGFR3 is considered as an inducer of lymphangiogenesis.86 By activating Smad4/TGFβ/VEGFC pathway, high level of miR27a accelerates lymphatic tube formation and lymphatic metastasis.90
miRNAs and Anoikis
Cancer generates mobile cells termed CTCs that immigrate from the primary environment and seep into the circulatory system.91 A type of programmed cell death termed anoikis is triggered when they leave the primary niche. CTCs must acquire the ability to resist anoikis and survive in the new background. Some malignant cells are equipped with the ability to resist anoikis or are less sensitive to anoikis.92 Itgα3 signaling pathway has been demonstrated to be critical in regulating sensitivity of colorectal CTCs to anoikis. High miR124 levels activate caspase 3 and induce anoikis by reducing the expression of Itgα3. High levels of Itgα3 or reduced miR124 indicate poor CRC prognosis.93 The expression of miR10a is upregulated in primary CRC tissues in SW480 cells compared to metastatic cancer tissue with lymph-node metastasis in SW620 cells. High miR10a levels induce cell mobility and decrease cell adhesion in primary CRC cells. When CRC cells detach from the primary niche, the expression of miR10a in CRC CTCs is decreased. Low miR10a levels enhance cell adhesion and promote anoikis resistance by increasing the expression of integrin and BCL2.94 Tumor immunoescape refers to TCs escaping from immune-system attack and surviving in metastasis cascades. High PDL1 levels on the tumor surface can bind to PD1 on T lymphocytes, leading to immunosuppression and contributing to tumor survival.95 The endogenous tumor suppressor miR93-5p directly suppresses the expression of PDL1. miR93-5p in CRC cells contributes to inhibition of immunoescape by inhibiting PDL1–PD1 interaction and improving the production of anti-inflammatory factors. High expression of PDL1 or low levels of miR93-5p indicate more CRC-metastasis potential.96
miRNAs and PMN
Cancer metastasis is organ- and tissue-specific rather than random, according to the seed–soil theory and PMN theory, and the site of metastasis is related to the integrin type on the cell surface.97 The PMN theory refers to primary cancer promoting a suitable microenvironment in distant organs or tissue for cancer-cells settlement.98 As for CRC, the liver and peritoneum are the two commonest sites of metastasis. Studies have found that primary CRC cells can facilitate PMN formation in the liver by releasing various miRNA-contained exosomes.4
miR21-contained exosomes are released from primary CRC cells and identified by macrophages in the liver, then miR21 stimulates polarization of macrophages and enhances the production of IL6. The secreted IL6 in turn enhances miR21 recruitment, which further expands the inflammatory response in the liver and consequently forms an inflammatory PMN site to facilitate liver metastasis for CRC.99 Similarly, the expression of miR25-3p, miR130b-3p, and miR425-5p stimulated by CXCL12 and CXCR4 is upregulated in CRC cells. When these miRNAs are secreted by CRC cells in exosomes and taken up by TAMs, they enhance the infiltration of TAMs and induce an inflammatory environment in the liver via inhibiting PTEN and activating the PI3K–Akt pathway.100
The reverse process of EMT, MET is also essential for metastasis. EMT allows emigration from primary tumor tissues. The MET process shifts the cancer cells to a less aggressive phenotype and allows cancer cells to settle in a distant location to go on to further proliferation.101 miR200c also expresses contrastingly between primary and metastatic tissue. The level of miR200c in primary tissue is lower than in metastatic tissue. Lowly expressed miR200c triggers EMT in the invasive margin of primary CRC tissue. When cells reach a suitable distant metastatic location, such as lymph nodes or the liver, the expression of miR200c is increased. High levels of miR200c promote MET and allow CRC cells to regain epithelial features and decrease mobility.102 Collectively, EMT triggered by low levels of miR200c in primary CRC tissue allows cancer-cell emigration and MET process allows the settlement of CTCs in distant tissues.
miRNAs and Stemness
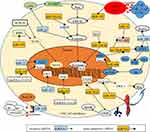
CSCs are subpopulation cancer-cell clusters that express specific markers, including CD44, CD24, CD133, LGR5, Pgp, and ABCG2. CSCs are equipped with abilities of self-renewal, multilineage-differentiation, tumorsphere formation, and conventional treatment resistance. A small group of CSCs can be activated and form complete tumor tissue. They are also considered the seeds of metastasis and treatment resistance.103 CRC-derived miRNAsregulate the stemness of CSCs via various signaling pathways (Figure 2).104 CSCs undergo asymmetric division to retain stem traits. This process produces partially differentiated cells and SCs.13 Epigenetically silenced miR34a accelerates the asymmetric division process of CRC SCs, enhances self-renewal ability, and increases the expression of stem markers by activating Notch and the Wnt–β-catenin pathway.105
The reverse process of differentiation happens in cancer when differentiated cancer cells reacquire stemness properties. CSCs interact with differentiated cancer cells and endow them with SC-like traits. Specifically, parent CSCs release exosomal miR146a to differentiated CRC cells and promote them to regain SC-like properties. The activated Wnt–β-catenin–TCF4–Rab27B pathway enhances the secretion of miR146a-encapsulated exosomes, which increases the expression of stem markers and self-renewal ability in recipient CRC cells.106 Stem markers mainly help to distinguish SCs and non-SCs. Some markers also contribute to conventional drug resistance, eg, Pgp, ABCG2, and CD44. CD44 is a stem marker, and high levels result in attenuated sensitivity to cetuximab. Ectopic expression of miR302 accelerates drug efflux in CRC chemotherapy by increasing the expression of CD44.107
miRNAs, Tumor Markers, and Treatment Resistance
miRNA expression in cancer patients is correlated with tumor burden, and dysregulation of miRNAs in cancer patients can act as a clinical diagnostic biomarker. They are detectable in blood, and levels correspond with the progression of cancer (Table 3). For example, levels of miR183-5p, miR365a-3p, and miR302a in CRC patients have been shown to be related to tumor size, extent of lymph-node metastasis, and whether distant metastasis has occurred.121 For a lack of symptoms in early stages, CRC patients often miss the opportunity for early diagnosis. Also, in the process ofpostoperative tumor review, miRNAs may serve as an important supplementary examination method to provide new detection methods for early diagnosis and postoperative review (Table 3).
 |
Table 2 Effects of miRNAs on the regulation of CRC stemness |
 |
Table 3 Effects of miRNAs in clinical applications |
Treatment resistance of malignant tumors often indicates poor prognosis, and the unregulated expression of some miRNAs is directly related to chemotherapy resistance. In certain cell lines with stem markers highly expressed, there is abnormal expression of miRNAs associated with resistance to fluorochemical and platinum-based chemotherapy.122 These miRNAs are related to drug efflux, DNA-damage repair, and drug metabolism. For example, thymidylate synthase is one of the metabolic enzymes of 5FU, and miR375-3p enhances the sensitivity of CRC cells to 5FU by regulating the expression of thymidylate synthase.123 Downregulation of miR4454 is related to drug resistance of CRC. Artificial induction of miR4454 expression leads to reduction in tumor tissue and increases sensitivity to 5FU, CPT11, and oxaliplatin.124 A drug transporter, MRP5 decreases sensitivity to oxaliplatin by increasing drug efflux and reducing intracellular oxaliplatin accumulation. miR128-3p is a direct inhibitor of MRP5. Expression of miR128-3p is reduced in oxaliplatin-resistant cell lines (HCT116OxR and HT29OxR). Increasing the level of miR128-3p can improve sensitivity to oxaliplatin by suppressing the expression of MRP5.125
Conclusion
CRC is the third-commonest cancer worldwide and metastasis the main cause of cancer-related death. Various proteins, lipids, and nucleotides including DNA and RNA regulate CRC metastasis in different aspects. Increasing evidence demonstrates that miRNAs participate in cancer metastasis at the epigenetic level. As a member of the ncRNA family, miRNAs have been found to participate in occurrence and development of CRC. miRNAs suppress posttranslational expression of metastasis-related genes or cause degradation of various mRNAs through an miRNA–mRNA interference model.
Firstly, miRNAs induce reprogramming of CRC cells to acquire mobility and generate mobile cells. Different miRNAs accelerate morphological changes in CRC cells and detachment from the ECM by affecting cytoskeleton remodeling, adhesion, and metabolism. miRNAs mediate the expression of EMT-related proteins (E-cadherin, N-cadherin, vimentin, and fibronectin) in CRC cells to attenuate polarity and acquire an aggressive phenotype during EMT–MET conversion.138 During this process, the deformed CRC cells accelerate degradation of the ECM and interact with other noncancer cells in the TME to promote invasion and migration. Secondly, miRNAs regulate angiogenesis, destroy endothelial barriers, and increase vascular permeability in primary CRC tissue by regulating several angiogenesis-associated factors (Ang2, HIF1, TGFβ, and VEGF), leading to the leakage of CRC cells into the bloodstream and the formation of CTCs. Thirdly, miRNAs endow CRC CTCs with anoikis resistance and facilitate their survival in the bloodstream. miRNAs regulate the expression of PDL1 on the CRC cell surface and free CRC cells from immune harm. Then, miRNAs-contained exosomes from primary CRC can educate distant sites to establish a favorable PMN and facilitate CTCs to settle there, and the miRNAs induce angiogenesis, MET, and inflammatory reactions in the distant site for further settlement. Finally, miRNAs maintain CRC stemness. CSCs are considered the seeds of metastasis and recurrence. Conventional treatment fails to eliminate CSCs completely, which leaves the potential for metastasis. There are special clusters in CRC with specific makers expressed on the cell surface (CD44, CD24, CD133, and LGR5), and they are long-lived in the body, with self-renewal ability, great cell populations, tumor initiation, and multilineage-differentiation potential.139 Clinically, miRNAs may be novel makers for early diagnosis and indicators for prognosis. They may also be potential methods to overcome CRC-treatment resistance.
We found that the level of miRNAs is in a dynamic state and has dual effects in CRC.miRNA levels in CRC alter in response to different microenvironments. They are regulated by various factors, such as TGFβ, EGFs, hypoxia, hyperglycemia, and inflammatory factors.Expression of miRNAs is affected by cell state. For example, the level of miR200c and miR10a differs in primary and metastatic CRC cells. miRNAs exist in various forms. miRNAs exist intracellularly and regulate the expression of various genes. In addition, miRNAs can also be secreted by CRC cells via lipid carriers, such as exosomes or protein carriers. miRNAs can work as oncogenic or tumor-suppressive factors in CRC progression (Tables 1 and 2).
Though the genesis of CSCs is unknown, retention of self-renewal ability and stem-related markers are upregulated when cells undergo EMT. Some studies have regarded the stemness as a consequence of EMT and miRNAs regulating EMT and stem properties synchronously.140,141 In normal tissue, with different stages and different types of tumors, patterns of miRNA expression exhibit different features. Firstly, expression profiles of miRNAs are different between the normal and tumor tissues. Secondly, miRNA expression shows tissue-specificity. Thirdly, expression profiles of miRNAs show significant difference among different cancers.142 There have been studies showing that expression differs between poorly differentiated and well-differentiated tumor tissue. In metastatic cancers with unknown primary origins or poorly differentiated malignant tumors, specific miRNA-expression profiles may help to determine tumor origin, including in CRC.143,144 The corresponding relationship between tumor stages and miRNA-expression levels ensures the possibility of miRNAs working as indicators for CRC prognosis. miRNA-expression profiles are informative, reflecting the developmental lineages and differentiation states of the tumors. A study has demonstrated that 48 miRNA markers encode 22 tissue types, with accuracy reaching 90%.143
Expression profiles of miRNAs differ among species. At the cellular level, they are regulated in nuclei and cytoplasm. The expression process of miRNAs includes miRNA transcription, procession by Drosha and Dicer, and then the most miRNAs mature in cytoplasm. Transcription factors, such as p53, Myc, ZEB1, and ZEB2, positively or negatively regulate the expression of miRNAs.145 The transcription miR34a is regulated by p53 in the nucleus.42 Drosha forms a complex called a microprocessor that determines the specificity of miRNAs. The level of Dicer also determines the level of miRNAs. Expression of Dicer increases as the density of cancer cells increases, which leads to increased expression of miR590-5p (density-sensitive miRNA).146
Cell–cell communication is a foundation of cellular biological phenomena. Differently from various signaling molecules, miRNAs act as signaling molecules and shuttle between CRC cells and other noncancer cells, providing new insight.147 miRNAs in the TME mediate signal exchange between cancer cells and noncancer cells and differentiated and undifferentiated cancer cells.148,149 They are even secreted into the bloodstreams by exosome, and can function in distant sites. In terms of chemotherapy resistance, fluorine and platinum drugs are first-line chemotherapeutics in CRC treatment. The dysregulation of miRNAs in CRC, such as in miR141, Let7, and miR375-3p, are involved in conventional chemotherapy resistance and poor survival. Levels of miRNAs in serum are related to CRC stage, and as biomarkers, miRNAs can be critical diagnostic and metastasis indicators. The quantitative detection of conservational miRNA markers may provide the possibility of detecting CRC recurrence and metastasis.150,151 It has been found that by artificially regulating the expression of miRNA in cancer cell, delivering miRNAs or their regulatory molecules to cancer cells can affect the rate of cancer progression. For example, artificial intracellular miR21 inhibitors can significantly increase the sensitivity of CRC cells to chemotherapy.152 In conclusion, reviewing the role of different miRNAs in CRC metastasis could benefit us tremendously in the field of CRC research and clinical therapy.
Abbreviations
5FU, 5-fluorouracil; Cdh, cadherin; Cld, claudin; CPT11, irinotecan; CXCR, chemokine receptor; Itgα, integrin α; Itgβ, integrin β; OS, overall survival; PFS, progression-free survival; TCF, T-cell factor.
Funding
This work was supported by the Natural Science Foundation of Jiangxi Province, China.
Disclosure
The authors declare no conflicts of interest for this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin. 2019;69(5):363–385. doi:10.3322/caac.21565
3. Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2019: a review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2019;69(3):184–210. doi:10.3322/caac.21557
4. Pretzsch E, Bosch F, Neumann J, et al. Mechanisms of metastasis in colorectal cancer and metastatic organotropism: hematogenous versus peritoneal spread. J Oncol. 2019;2019:7407190. doi:10.1155/2019/7407190
5. Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi:10.1016/j.tcb.2018.12.001
6. Yuzhalin AE, Lim SY, Kutikhin AG, Gordon-Weeks AN. Dynamic matrisome: ECM remodeling factors licensing cancer progression and metastasis. Biochim Biophys Acta Rev Cancer. 2018;1870(2):207–228. doi:10.1016/j.bbcan.2018.09.002
7. Soheilifar MH, Grusch M, Neghab HK, et al. Angioregulatory microRNAs in colorectal cancer. Cancers (Basel). 2019;12:1. doi:10.3390/cancers12010071
8. Zeng Z, Li Y, Pan Y, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. doi:10.1038/s41467-018-07810-w
9. Stacker SA, Achen MG, Jussila L, Baldwin ME, Alitalo K. Lymphangiogenesis and cancer metastasis. Nat Rev Cancer. 2002;2(8):573–583. doi:10.1038/nrc863
10. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. CELL. 2017;168(4):670–691. doi:10.1016/j.cell.2016.11.037
11. Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct Target Ther. 2020;5:28.
12. Quintero-Fabian S, Arreola R, Becerril-Villanueva E, et al. Role of matrix metalloproteinases in angiogenesis and cancer. Front Oncol. 2019;9:1370. doi:10.3389/fonc.2019.01370
13. Nandy SB, Lakshmanaswamy R. Cancer stem cells and metastasis. Prog Mol Biol Transl Sci. 2017;151:137–176.
14. Raza U, Zhang JD, Sahin O. MicroRNAs: master regulators of drug resistance, stemness, and metastasis. J Mol Med (Berl). 2014;92(4):321–336. doi:10.1007/s00109-014-1129-2
15. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
16. Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. NATURE. 2008;455(7209):64–71. doi:10.1038/nature07242
17. Garofalo M, Croce CM. microRNAs: master regulators as potential therapeutics in cancer. Annu Rev Pharmacol Toxicol. 2011;51:25–43. doi:10.1146/annurev-pharmtox-010510-100517
18. Trepat X, Chen Z, Jacobson K. Cell migration. Compr Physiol. 2012;2(4):2369–2392.
19. Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171(24):5507–5523. doi:10.1111/bph.12704
20. Wu JI, Wang LH. Emerging roles of gap junction proteins connexins in cancer metastasis, chemoresistance and clinical application. J Biomed Sci. 2019;26(1):8.
21. Wilson K, Lewalle A, Fritzsche M, Thorogate R, Duke T, Charras G. Mechanisms of leading edge protrusion in interstitial migration. NAT COMMUN. 2013;4:2896. doi:10.1038/ncomms3896
22. Kim CW, Oh ET, Kim JM, et al. Corrigendum to “Hypoxia-induced microRNA-590-5p promotes colorectal cancer progression by modulating matrix metalloproteinase activity” [Cancer Lett. 416 (2018) 31-41]. Cancer Lett. 2019;455::73. doi:10.1016/j.canlet.2019.04.024
23. Julian L, Olson MF. Rho-associated coiled-coil containing kinases (ROCK): structure, regulation, and functions. Small GTPases. 2014;5:e29846. doi:10.4161/sgtp.29846
24. Yu X, Wang D, Wang X, et al. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J Exp Clin Cancer Res. 2019;38(1):32. doi:10.1186/s13046-018-1014-x
25. Zhuang M, Zhao S, Jiang Z, et al. MALAT1 sponges miR-106b-5p to promote the invasion and metastasis of colorectal cancer via SLAIN2 enhanced microtubules mobility. EBIOMEDICINE. 2019;41:286–298. doi:10.1016/j.ebiom.2018.12.049
26. Liu H, Liu Y, Sun P, et al. Bai J and Cui B. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci (Lond). 2020;134(4):419–434. doi:10.1042/CS20191087
27. Sheng N, Tan G, You W, et al. MiR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017;6(6):1331–1340. doi:10.1002/cam4.1029
28. Wei AW, Li LF. Long non-coding RNA SOX21-AS1 sponges miR-145 to promote the tumorigenesis of colorectal cancer by targeting MYO6. Biomed Pharmacother. 2017;96:953–959. doi:10.1016/j.biopha.2017.11.145
29. Anderson LR, Owens TW, Naylor MJ. Integrins in development and cancer. Biophys Rev. 2014;6(2):191–202.
30. Barkan D, Chambers AF. beta1-integrin: a potential therapeutic target in the battle against cancer recurrence. Clin Cancer Res. 2011;17(23):7219–7223. doi:10.1158/1078-0432.CCR-11-0642
31. Yang X, Chen J, Liao Y, et al. MiR-27b-3p promotes migration and invasion in colorectal cancer cells by targeting HOXA10. Biosci Rep. 2019;39(12).
32. Zhao Y, Miao G, Li Y, et al. MicroRNA- 130b suppresses migration and invasion of colorectal cancer cells through downregulation of integrin beta1 [corrected]. PLoS One. 2014;9(2):e87938.
33. Laudato S, Patil N, Abba ML, et al. 53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int J Cancer. 2017;141(9):1879–1890. doi:10.1002/ijc.30854
34. Huang L, Cai JL, Huang PZ, et al. miR19b-3p promotes the growth and metastasis of colorectal cancer via directly targeting ITGB8. Am J Cancer Res. 2017;7(10):1996–2008.
35. Yoo HI, Kim BK, Yoon SK. MicroRNA-330-5p negatively regulates ITGA5 expression in human colorectal cancer. Oncol Rep. 2016;36(5):3023–3029. doi:10.3892/or.2016.5092
36. Liberti MV, Locasale JW. Correction to: ‘The Warburg Effect: how does it benefit cancer cells?’: [Trends in Biochemical Sciences, 41 (2016) 211]. Trends Biochem Sci. 2016;41(3):287. doi:10.1016/j.tibs.2016.01.004
37. Barisciano G, Colangelo T, Rosato V, et al. Correction: miR-27a is a master regulator of metabolic reprogramming and chemoresistance in colorectal cancer. Br J Cancer. 2020.
38. Deng F, Zhou R, Lin C, et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics. 2019;9(4):1001–1014. doi:10.7150/thno.30056
39. Wang X, Zhang H, Yang H, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14(3):539–555. doi:10.1002/1878-0261.12629
40. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi:10.1038/s41580-018-0080-4
41. Vu T, Datta PK. Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers (Basel). 2017;9:12. doi:10.3390/cancers9120171
42. Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic Effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology. 2017;153(2):505–520. doi:10.1053/j.gastro.2017.04.017
43. Liu Y, Gu Y, Cao X. The exosomes in tumor immunity. Oncoimmunology. 2015;4(9):e1027472.
44. Ruiz-Lopez L, Blancas I, Garrido JM, et al. The role of exosomes on colorectal cancer: a review. J Gastroenterol Hepatol. 2018;33(4):792–799. doi:10.1111/jgh.14049
45. Sanchez-Martin D, Otsuka A, Kabashima K, et al. Effects of DLC1 deficiency on endothelial cell contact growth inhibition and angiosarcoma progression. J Natl Cancer Inst. 2018;110(4):390–399. doi:10.1093/jnci/djx219
46. Sun Z, Ou C, Liu J, et al. YAP1-induced MALAT1 promotes epithelial-mesenchymal transition and angiogenesis by sponging miR-126-5p in colorectal cancer. Oncogene. 2019;38(14):2627–2644. doi:10.1038/s41388-018-0628-y
47. Ren LL, Yan TT, Shen CQ, et al. The distinct role of strand-specific miR-514b-3p and miR-514b-5p in colorectal cancer metastasis. Cell Death Dis. 2018;9(6):687. doi:10.1038/s41419-018-0732-5
48. Xie Y, Zhao J, Liang Y, et al. MicroRNA-10b controls the metastasis and proliferation of colorectal cancer cells by regulating Kruppel-like factor 4. Artif Cells Nanomed Biotechnol. 2019;47(1):1722–1729. doi:10.1080/21691401.2019.1606006
49. Tang W, Zhu Y, Gao J, et al. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br J Cancer. 2014;110(2):450–458. doi:10.1038/bjc.2013.724
50. Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. LncRNA H19/miR-29b-3p/PGRN axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on wnt signaling. Mol Cells. 2018;41(5):423–435.
51. Li T, Jian X, He H, et al. MiR-452 promotes an aggressive colorectal cancer phenotype by regulating a Wnt/beta-catenin positive feedback loop. J Exp Clin Cancer Res. 2018;37(1):238. doi:10.1186/s13046-018-0879-z
52. Bu P, Wang L, Chen KY, et al. miR-1269 promotes metastasis and forms a positive feedback loop with TGF-beta. Nat Commun. 2015;6:6879. doi:10.1038/ncomms7879
53. Zhao S, Sun H, Jiang W, et al. miR-4775 promotes colorectal cancer invasion and metastasis via the Smad7/TGFbeta-mediated epithelial to mesenchymal transition. Mol Cancer. 2017;16(1):12. doi:10.1186/s12943-017-0585-z
54. Cai HK, Chen X, Tang YH, Deng YC. MicroRNA-194 modulates epithelial-mesenchymal transition in human colorectal cancer metastasis. Onco Targets Ther. 2017;10:1269–1278. doi:10.2147/OTT.S125172
55. Sun J, Hu J, Wang G, et al. LncRNA TUG1 promoted KIAA1199 expression via miR-600 to accelerate cell metastasis and epithelial-mesenchymal transition in colorectal cancer. J Exp Clin Cancer Res. 2018;37(1):106. doi:10.1186/s13046-018-0771-x
56. Shen Z, Zhou R, Liu C, et al. MicroRNA-105 is involved in TNF-alpha-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8(12):3213. doi:10.1038/s41419-017-0048-x
57. Shen T, Cheng X, Liu X, et al. Circ_0026344 restrains metastasis of human colorectal cancer cells via miR-183. Artif Cells Nanomed Biotechnol. 2019;47(1):4038–4045. doi:10.1080/21691401.2019.1669620
58. Geng Y, Zheng X, Hu W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond). 2019;133(10):1197–1213. doi:10.1042/CS20190286
59. Wang H, Yan B, Zhang P, et al. MiR-496 promotes migration and epithelial-mesenchymal transition by targeting RASSF6 in colorectal cancer. J Cell Physiol. 2020;235(2):1469–1479. doi:10.1002/jcp.29066
60. Tang W, Hong L, Dai W, et al. MicroRNA500a5p inhibits colorectal cancer cell invasion and epithelialmesenchymal transition. INT J ONCOL. 2020.
61. Li J, Peng W, Yang P, et al. MicroRNA-1224-5p inhibits metastasis and epithelial-mesenchymal transition in colorectal cancer by targeting SP1-Mediated NF-kappaB signaling pathways. FRONT ONCOL. 2020;10:294. doi:10.3389/fonc.2020.00294
62. Karimi DF, Amini R, Saidijam M, Najafi R. miR-200c, a tumor suppressor that modulate the expression of cancer stem cells markers and epithelial-mesenchymal transition in colorectal cancer. J Cell Biochem. 2018;119(7):6288–6295. doi:10.1002/jcb.26880
63. Chen H, Xiao Q, Hu Y, et al. ANGPTL1 attenuates colorectal cancer metastasis by up-regulating microRNA-138. J Exp Clin Cancer Res. 2017;36(1):78. doi:10.1186/s13046-017-0548-7
64. Shen X, Jiang H, Chen Z, et al. MicroRNA-145 inhibits cell migration and invasion in colorectal cancer by targeting TWIST. Onco Targets Ther. 2019;12:10799–10809. doi:10.2147/OTT.S216147
65. Chen J, Zhang H, Chen Y, et al. miR-598 inhibits metastasis in colorectal cancer by suppressing JAG1/Notch2 pathway stimulating EMT. Exp Cell Res. 2017;352(1):104–112. doi:10.1016/j.yexcr.2017.01.022
66. Wang YB, Shi Q, Li G, Zheng JH, Lin J, Qiu W. MicroRNA-488 inhibits progression of colorectal cancer via inhibition of the mitogen-activated protein kinase pathway by targeting claudin-2. Am J Physiol Cell Physiol. 2019;316(1):C33–C47. doi:10.1152/ajpcell.00047.2018
67. Chen YC, Ou MC, Fang CW, Lee TH, Tzeng SL. High glucose concentrations negatively regulate the IGF1R/Src/ERK Axis through the MicroRNA-9 in colorectal cancer. Cells-Basel. 2019;8:4.
68. Karimi L, Zeinali T, Hosseinahli N, et al. miRNA-143 replacement therapy harnesses the proliferation and migration of colorectal cancer cells in vitro. J Cell Physiol. 2019;234(11):21359–21368. doi:10.1002/jcp.28745
69. Ma W, Liu B, Li J, et al. MicroRNA-302c represses epithelial-mesenchymal transition and metastasis by targeting transcription factor AP-4 in colorectal cancer. Biomed Pharmacother. 2018;105:670–676. doi:10.1016/j.biopha.2018.06.025
70. Lun W, Wu X, Deng Q, Zhi F. MiR-218 regulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via targeting CTGF. Cancer Cell Int. 2018;18:83. doi:10.1186/s12935-018-0575-2
71. Park YR, Seo SY, Kim SL, et al. MiRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targetting TM4SF1. Biosci Rep. 2018;38:5. doi:10.1042/BSR20180664
72. Chen X, Zeng K, Xu M, et al. 53-induced miR-1249 inhibits tumor growth, metastasis, and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis. 2019;10(2):131. doi:10.1038/s41419-018-1188-3
73. Li J, Zhao LM, Zhang C, et al. The lncRNA FEZF1-AS1 promotes the progression of colorectal cancer through regulating OTX1 and targeting miR-30a-5p. Oncol Res. 2020;28(1):51–63. doi:10.3727/096504019X15619783964700
74. Park YR, Kim SL, Lee MR, et al. MicroRNA-30a-5p (miR-30a) regulates cell motility and EMT by directly targeting oncogenic TM4SF1 in colorectal cancer. J Cancer Res Clin Oncol. 2017;143(10):1915–1927. doi:10.1007/s00432-017-2440-4
75. Du F, Cao T, Xie H, et al. KRAS Mutation-Responsive miR-139-5p inhibits colorectal cancer progression and is repressed by Wnt signaling. Theranostics. 2020;10(16):7335–7350. doi:10.7150/thno.45971
76. Lin X, Wang S, Sun M, et al. Correction to: miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 2019;12(1):122. doi:10.1186/s13045-019-0810-x
77. Wei C, Yang C, Wang S, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18(1):64. doi:10.1186/s12943-019-0976-4
78. Fan L, Wu Y, Wang J. Sevoflurane inhibits the migration and invasion of colorectal cancer cells through regulating ERK/MMP-9 pathway by up-regulating miR-203. Eur J Pharmacol. 2019;850:43–52. doi:10.1016/j.ejphar.2019.01.025
79. Zhang L, Dong Y, Zhu N, et al. microRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. MOL CANCER. 2014;13:124.
80. Yu L, Lu Y, Han X, et al. microRNA −140-5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem Cell Res Ther. 2016;7(1):180. doi:10.1186/s13287-016-0438-5
81. Hu JL, Wang W, Lan XL, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18(1):91. doi:10.1186/s12943-019-1019-x
82. Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. THERANOSTICS. 2018;8(14):3932–3948. doi:10.7150/thno.25541
83. Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. ANGIOGENESIS. 2017;20(4):409–426. doi:10.1007/s10456-017-9562-9
84. Xu Z, Zhu C, Chen C, et al. CCL19 suppresses angiogenesis through promoting miR-206 and inhibiting Met/ERK/Elk-1/HIF-1alpha/VEGF-A pathway in colorectal cancer. Cell Death Dis. 2018;9(10):974.
85. Yamakuchi M, Lotterman CD, Bao C, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107(14):6334–6339. doi:10.1073/pnas.0911082107
86. Melincovici CS, Bosca AB, Susman S, et al. Vascular endothelial growth factor (VEGF) - key factor in normal and pathological angiogenesis. Rom J Morphol Embryol. 2018;59(2):455–467.
87. Hu HY, Yu CH, Zhang HH, et al. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470–477.
88. Guan H, Xie L, Leithauser F, et al. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116(9):1469–1478. doi:10.1182/blood-2009-12-256446
89. Yamada NO, Heishima K, Akao Y, Senda T. Extracellular vesicles containing MicroRNA-92a-3p facilitate partial endothelial-mesenchymal transition and angiogenesis in endothelial cells. Int J Mol Sci. 2019;20:18. doi:10.3390/ijms20184406
90. Xu Q, Tong JL, Zhang CP, Xiao Q, Lin XL, Xiao XY. miR-27a induced by colon cancer cells in HLECs promotes lymphangiogenesis by targeting SMAD4. PLoS One. 2017;12(10):e186718.
91. Fabisiewicz A, Grzybowska E. CTC clusters in cancer progression and metastasis. Med Oncol. 2017;34(1):12. doi:10.1007/s12032-016-0875-0
92. Simpson CD, Anyiwe K, Schimmer AD. Anoikis resistance and tumor metastasis. Cancer Lett. 2008;272(2):177–185. doi:10.1016/j.canlet.2008.05.029
93. Sa KD, Zhang X, Li XF, et al. A miR-124/ITGA3 axis contributes to colorectal cancer metastasis by regulating anoikis susceptibility. Biochem Biophys Res Commun. 2018;501(3):758–764. doi:10.1016/j.bbrc.2018.05.062
94. Liu Y, Zhang Y, Wu H, et al. Mir-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017;8(4):e2739. doi:10.1038/cddis.2017.61
95. Carter L, Fouser LA, Jussif J, et al. PD-1:PD-Linhibitory pathway affects both CD4(+) and CD8(+) T cells and is overcome by IL-2. Eur J Immunol. 2002;32(3):634–643. doi:10.1002/1521-4141(200203)32:3<634::AID-IMMU634>3.0.CO;2-9
96. Chen YL, Wang GX, Lin BA, Huang JS. MicroRNA-93-5p expression in tumor tissue and its tumor suppressor function via targeting programmed death ligand-1 in colorectal cancer. Cell Biol Int. 2020;44(5):1224–1236. doi:10.1002/cbin.11323
97. Peinado H, Zhang H, Matei IR, et al. Pre-metastatic niches: organ-specific homes for metastases. Nat Rev Cancer. 2017;17(5):302–317.
98. Paget S. The distribution of secondary growths in cancer of the breast. 1889 Cancer Metastasis Rev. 1989;8(2):98–101.
99. Shao Y, Chen T, Zheng X, et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis. 2018;39(11):1368–1379. doi:10.1093/carcin/bgy115
100. Wang D, Wang X, Si M, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi:10.1016/j.canlet.2020.01.005
101. Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9(4):265–273. doi:10.1038/nrc2620
102. Hur K, Toiyama Y, Takahashi M, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62(9):1315–1326. doi:10.1136/gutjnl-2011-301846
103. Adorno-Cruz V, Kibria G, Liu X, et al. Cancer stem cells: targeting the roots of cancer, seeds of metastasis, and sources of therapy resistance. Cancer Res. 2015;75(6):924–929. doi:10.1158/0008-5472.CAN-14-3225
104. Jiao X, Qian X, Wu L, et al. microRNA: the impact on cancer stemness and therapeutic resistance. Cells-Basel. 2019;9:1.
105. Wang L, Bu P, Ai Y, et al. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife. 2016;5.
106. Cheng WC, Liao TT, Lin CC, et al. RAB27B-activated secretion of stem-like tumor exosomes delivers the biomarker microRNA-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer. 2019;145(8):2209–2224. doi:10.1002/ijc.32338
107. Sun L, Fang Y, Wang X, et al. miR-302a inhibits metastasis and cetuximab resistance in colorectal cancer by targeting NFIB and CD44. Theranostics. 2019;9(26):8409–8425. doi:10.7150/thno.36605
108. Fan D, Lin X, Zhang F, et al. MicroRNA 26b promotes colorectal cancer metastasis by downregulating phosphatase and tensin homolog and wingless-type MMTV integration site family member 5A. Cancer Sci. 2018;109(2):354–362. doi:10.1111/cas.13451
109. Wang LQ, Yu P, Li B, et al. miR-372 and miR-373 enhance the stemness of colorectal cancer cells by repressing differentiation signaling pathways. Mol Oncol. 2018;12(11):1949–1964. doi:10.1002/1878-0261.12376
110. Mukohyama J, Isobe T, Hu Q, et al. miR-221 targets QKI to enhance the tumorigenic capacity of human colorectal cancer stem cells. Cancer Res. 2019;79(20):5151–5158. doi:10.1158/0008-5472.CAN-18-3544
111. Yu Y, Kanwar SS, Patel BB, et al. MicroRNA-21 induces stemness by downregulating transforming growth factor beta receptor 2 (TGFbetaR2) in colon cancer cells. Carcinogenesis. 2012;33(1):68–76. doi:10.1093/carcin/bgr246
112. Lai HT, Tseng WK, Huang SW, Chao TC, Su Y. MicroRNA-203 diminishes the stemness of human colon cancer cells by suppressing GATA6 expression. J Cell Physiol. 2020;235(3):2866–2880. doi:10.1002/jcp.29192
113. Jiang S, Miao D, Wang M, Lv J, Wang Y, Tong J. MiR-30-5p suppresses cell chemoresistance and stemness in colorectal cancer through USP22/Wnt/beta-catenin signaling axis. J Cell Mol Med. 2019;23(1):630–640. doi:10.1111/jcmm.13968
114. Ma X, Liu J, Li J, et al. miR-139-5p reverses stemness maintenance and metastasis of colon cancer stem-like cells by targeting E2-2. J Cell Physiol. 2019;234(12):22703–22718. doi:10.1002/jcp.28836
115. Jin Y, Wang M, Hu H, Huang Q, Chen Y, Wang G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int J Biol Macromol. 2018;117:445–453. doi:10.1016/j.ijbiomac.2018.05.151
116. Yu Y, Nangia-Makker P, Farhana L, Majumdar A. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer. 2017;16(1):155. doi:10.1186/s12943-017-0725-5
117. Tang D, Yang Z, Long F, et al. Long noncoding RNA MALAT1 mediates stem cell-like properties in human colorectal cancer cells by regulating miR-20b-5p/Oct4 axis. J Cell Physiol. 2019;234(11):20816–20828. doi:10.1002/jcp.28687
118. Chen DL, Lu YX, Zhang JX, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. THERANOSTICS. 2017;7(19):4836–4849. doi:10.7150/thno.20942
119. Khodadadi KA, Saidijam M, Amini R, Samadi P, Najafi R. Induction of let-7e gene expression attenuates oncogenic phenotype in HCT-116 colorectal cancer cells through targeting of DCLK1 regulation. Life Sci. 2019;228:221–227. doi:10.1016/j.lfs.2019.05.005
120. Ji D, Zhan T, Li M, et al. Enhancement of sensitivity to chemo/radiation therapy by using miR-15b against DCLK1 in colorectal cancer. Stem Cell Rep. 2018;11(6):1506–1522. doi:10.1016/j.stemcr.2018.10.015
121. Hong YG, Xin C, Zheng H, et al. miR-365a-3p regulates ADAM10-JAK-STAT signaling to suppress the growth and metastasis of colorectal cancer cells. J Cancer. 2020;11(12):3634–3644. doi:10.7150/jca.42731
122. Bukowski K, Kciuk M, Kontek R. Mechanisms of multidrug resistance in cancer chemotherapy. INT J MOL SCI. 2020;21:9. doi:10.3390/ijms21093233
123. Xu F, Ye ML, Zhang YP, et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020;111(5):1528–1541. doi:10.1111/cas.14356
124. Kannathasan T, Kuo WW, Chen MC, et al. Chemoresistance-associated silencing of miR-4454 promotes colorectal cancer aggression through the GNL3L and NF-kappaB pathway. Cancers (Basel). 2020;12:5. doi:10.3390/cancers12051231
125. Liu T, Zhang X, Du L, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18(1):43. doi:10.1186/s12943-019-0981-7
126. Tang Y, Zhao Y, Song X, Song X, Niu L, Xie L. Tumor-derived exosomal miRNA-320d as a biomarker for metastatic colorectal cancer. J Clin Lab Anal. 2019;33(9):e23004. doi:10.1002/jcla.23004
127. Sun Y, Yang B, Lin M, et al. Identification of serum miR-30a-5p as a diagnostic and prognostic biomarker in colorectal cancer. Cancer Biomark. 2019;24(3):299–305. doi:10.3233/CBM-182129
128. Sabry D, El-Deek S, Maher M, et al. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1alpha-VEGF signaling pathway. Mol Cell Biochem. 2019;454(1–2):177–189.
129. Choi HH, Cho YS, Choi JH, Kim HK, Kim SS, Chae HS. Stool-Based miR-92a and miR-144* as noninvasive biomarkers for colorectal cancer screening. Oncology. 2019;97(3):173–179. doi:10.1159/000500639
130. Ulivi P, Canale M, Passardi A, et al. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int J Mol Sci. 2018;19:1. doi:10.3390/ijms19010307
131. Zhang Y, Liu X, Zhang J, et al. Inhibition of miR-19a partially reversed the resistance of colorectal cancer to oxaliplatin via PTEN/PI3K/AKT pathway. Aging (Albany NY). 2020;12(7):5640–5650. doi:10.18632/aging.102929
132. Liang H, Xu Y, Zhang Q, et al. MiR-483-3p regulates oxaliplatin resistance by targeting FAM171B in human colorectal cancer cells. Artif Cells Nanomed Biotechnol. 2019;47(1):725–736. doi:10.1080/21691401.2019.1569530
133. Liu N, Li J, Zhao Z, et al. MicroRNA-302a enhances 5-fluorouracil-induced cell death in human colon cancer cells. Oncol Rep. 2017;37(1):631–639. doi:10.3892/or.2016.5237
134. Meng X, Fu R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. Onco Targets Ther. 2018;11:1757–1765. doi:10.2147/OTT.S159093
135. Hsu HH, Kuo WW, Shih HN, et al. FOXC1 regulation of miR-31-5p confers oxaliplatin resistance by targeting LATS2 in colorectal cancer. Cancers (Basel). 2019;11:10. doi:10.3390/cancers11101576
136. Wang M, Han D, Yuan Z, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(12):1149. doi:10.1038/s41419-018-1187-4
137. Yu X, Shi W, Zhang Y, et al. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci Rep. 2017;7:42226. doi:10.1038/srep42226
138. Brabletz T, Jung A, Reu S, et al. Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A. 2001;98(18):10356–10361. doi:10.1073/pnas.171610498
139. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi:10.1073/pnas.0530291100
140. Fabregat I, Malfettone A, Soukupova J. New insights into the crossroads between EMT and stemness in the context of cancer. J Clin Med. 2016;5:3. doi:10.3390/jcm5030037
141. Lan L, Luo Y, Cui D, et al. Epithelial-mesenchymal transition triggers cancer stem cell generation in human thyroid cancer cells. Int J Oncol. 2013;43(1):113–120. doi:10.3892/ijo.2013.1913
142. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi:10.1038/nature03702
143. Rosenfeld N, Aharonov R, Meiri E, et al. MicroRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–469. doi:10.1038/nbt1392
144. Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi:10.1073/pnas.0510565103
145. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524.
146. Ou C, Sun Z, Li X, et al. MiR-590-5p, a density-sensitive microRNA, inhibits tumorigenesis by targeting YAP1 in colorectal cancer. Cancer Lett. 2017;399:53–63. doi:10.1016/j.canlet.2017.04.011
147. Kogure A, Kosaka N, Ochiya T. Cross-talk between cancer cells and their neighbors via miRNA in extracellular vesicles: an emerging player in cancer metastasis. J Biomed Sci. 2019;26(1):7. doi:10.1186/s12929-019-0500-6
148. Yang F, Ning Z, Ma L, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. 2017;16(1):148.
149. Islam MS, Ciavattini A, Petraglia F, Castellucci M, Ciarmela P. Extracellular matrix in uterine leiomyoma pathogenesis: a potential target for future therapeutics. Hum Reprod Update. 2018;24(1):59–85.
150. Wang J, Ni J, Beretov J, Thompson J, Graham P, Li Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit Rev Oncol Hematol. 2020;145:102860. doi:10.1016/j.critrevonc.2019.102860
151. Joyce DP, Kerin MJ, Dwyer RM. Exosome-encapsulated microRNAs as circulating biomarkers for breast cancer. Int J Cancer. 2016;139(7):1443–1448. doi:10.1002/ijc.30179
152. Liang G, Zhu Y, Ali DJ, et al. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18(1):10.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.