Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
MicroRNAs as Novel Biomarkers for P2Y12 – Inhibitors Resistance Prediction
Authors Rytkin E , Mirzaev K, Bure I, Akmalova K , Abdullaev S, Kachanova A, Smirnov V, Grishina E, Lyakhova N, Aleshkovich E, Saribekian A, Andreev D , Shabunin A, Sychev D
Received 16 June 2021
Accepted for publication 24 November 2021
Published 2 December 2021 Volume 2021:14 Pages 1575—1582
DOI https://doi.org/10.2147/PGPM.S324612
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Martin H Bluth
Eric Rytkin,1 Karin Mirzaev,1 Irina Bure,1 Kristina Akmalova,1 Sherzod Abdullaev,1 Anastasiia Kachanova,1 Valery Smirnov,2,3 Elena Grishina,1 Natalia Lyakhova,4 Elena Aleshkovich,4 Anna Saribekian,2 Denis Andreev,2 Alexey Shabunin,4 Dmitry Sychev1
1Institute for Molecular and Personalized Medicine, Russian Medical Academy of Continuous Professional Education, Moscow, Russian Federation; 2Sechenov University, Moscow, Russian Federation; 3NRC Institute of Immunology FMBA of Russia, Moscow, Russian Federation; 4S.P. Botkin City Clinical Hospital, Moscow, Russian Federation
Correspondence: Eric Rytkin Email [email protected]
Aim: The aim of this study is to assess 6 micro-RNAs: miR-126, miR-223, miR-150, miR-29, miR-34, miR-142 as potential biomarkers for P2Y12- inhibitors resistance prediction.
Methods: Eighty patients with an acute coronary syndrome undergoing percutaneous coronary intervention treated in a multidisciplinary hospital in Moscow with DAPT (either with ticagrelor, n=45, or clopidogrel, n=35) were enrolled. The carriership of 6 clinically relevant polymorphisms for ticagrelor and 17 for clopidogrel was detected. Expression levels of six prospective miRNAs were measured. The activity of CYP3A4 isoenzyme was measured as the ratio of the concentrations of cortisol and 6β-hydroxycortisol.
Results: The polymorphisms of the P2Y12-inhibitors ADME genes that demonstrated statistically significant connection with miRNA expression levels are as follows: P2Y12R (A>G, rs3732759) and miR-29 (p=0.017), miR-34 (p=0.003); CYP2C19*17 (C-806T, rs1224856) and miR-142 (p=0.012); PON1 (Q192R, rs662) and miR-29 (p=0.004), ABCG2 (G>T, rs2231142) and miR-34 (p=0.007). MiRNAs expression levels showed connection with the results of the platelet reactivity assessment by utilizing VerifyNow assay (“Instrumentation laboratory”, MA, US). MiR-126 (β coefficient=− 0.076, SE=0.032, p=0.021), miR-223 (β coefficient=− 0.089, SE=0.041, p=0.032), miR-29 (β coefficient=− 0.042, SE=0.018, p=0.026), miR-142 (β coefficient=− 0.072, SE=0.026, p=0.008) have the potential to be used as biomarkers and may substitute platelet reactivity testing.
Conclusion: This study has revealed new biomarkers for P2Y12-inhibitors resistance testing: miR-29, miR-34, miR-126, miR-142, miR-223.
Keywords: biomarker, miRNA, polymorphism, acute coronary syndrome, pharmacogenomics
Plain Language Summary
- There is an unmet need for a novel biomarker for P2Y12 - Inhibitors Resistance Prediction
- The choice of an appropriate P2Y12-inhibitor as part of DAPT poses a challenge to cardiologists
- miR-29, miR-34, miR-126, miR-142 and miR-223 can be used as novel biomarkers
- These miRNAs may be included into future smart diagnostic tools
Introduction
Patients with an acute coronary syndrome [ACS] who undergo percutaneous coronary intervention [PCI] receive dual antiplatelet therapy [DAPT] as a standard of care.1 DAPT usually includes aspirin and a P2Y12-inhibitor. The DAPT therapy works towards prevention of thrombotic complications: cardiovascular death, myocardial infarction, stent thrombosis.2
Nowadays, 3 P2Y12-inhibitors are available per os: ticagrelor, clopidogrel, prasugrel. Clopidogrel is a prodrug, hence it needs to be metabolized in the liver, and there are some enzymes involved in its metabolism: CYP2C19, CYP2B6, CYP1A2, CYP3A4, CYP3A5, among them.3 Not only does metabolism of clopidogrel involve a two-stage transformation in the liver, but also transporter P-glycoproteins to facilitate prodrug absorption through the intestinal cells. ABCB1 gene is responsible for its regulation. All this contributes to a number of sites for a possible breakdown, which leads to altered response to clopidogrel. This high number of sites for a possible breakdown accounts for the number of patients who are resistant to clopidogrel, which can be as high as 35% of the population.4 As observed in a number of observational and randomized trials, a personalized approach to antiplatelet therapy may lead towards a decrease in the number of complications.5–8
Ticagrelor and prasugrel are considered more potent P2Y12–inhibitors in terms of the number of thrombotic complications among patients with an acute coronary syndrome undergoing percutaneous coronary intervention.9,10 Higher potency comes at a price of a higher number of bleeding complications, lower compliance due to high costs of the drugs. In addition, ticagrelor and prasugrel are not considered the drugs of choice when used as a combined antithrombotic treatment.11
The choice of an appropriate P2Y12-inhibitor to be used as part of DAPT poses a challenge to cardiologists as they need to know the patient’s characteristics to favour a certain drug.12
There were some trials which assessed doubling and tripling the dose of clopidogrel for low responders. However, the current strategy for low responders is switching to another P2Y12 inhibitor like ticagrelor or prasugrel.2,13–15
Various approaches are used for selection of an adequate antiplatelet therapy: genetic testing is implemented, and platelet reactivity is measured using the VerifyNow P2Y12 Assay.16–18 Currently, the Clinical Pharmacogenetics Implementation Consortium (CPIC) recommends genetic testing and genotype-directed treatment for high-risk patients.2 Therefore, CYP2C19 poor metabolizers should be prescribed an alternative antiplatelet regimen.19
Although, both genetic testing and platelet reactivity measurements are implemented, there is an unmet need for newer biomarkers developed for a simple, quick, reliable and comprehensive prediction of insufficient response to clopidogrel.20 And micro-RNAs are described in literature as such potential biomarkers.21–25
These micro-RNAs are small non-coding sequences of nucleotides which bind to mRNA sites and block transcription. This causes a decrease in the production of protein. The miRNA-mRNA mechanism is a fine instrument of gene expression regulation.26 The miRNAs may regulate ADME genes and affect the effective drug concentration in blood.
Micro-RNAs can be biomarkers for clopidogrel resistance through the ADME genes involved in the metabolism of clopidogrel.27
In this article, we suggest six prospective microRNAs as potential biomarkers for guiding antiplatelet therapy among patients with ACS who have undergone PCI.
Materials and Methods
Patients
Eighty patients with an acute coronary syndrome undergoing percutaneous coronary intervention treated in a multidisciplinary hospital in Moscow were consecutively enrolled.
The study was approved by the Ethics Committee of Russian Medical Academy of Continuous Professional Education, Moscow, Russian Federation and conducted in accordance with the Declaration of Helsinki; all the patients gave written informed consent for participation.
Inclusion criteria: presence of an informed written consent, age above 18 years, ACS event less than 7 days. Exclusion criteria: pregnancy, lactation, active internal bleeding, liver cirrhosis with C Child-Pugh liver insufficiency; chronic renal disease, HIV-infection, alcohol/drug addiction; mitral valve stenosis moderate and severe, mechanical heart valves, severe psychiatric disorders, allergic reactions/drug intolerance. The study assessed the secondary outcomes (рlatelet reactivity units, miRNAs expression levels). As part of dual antiplatelet therapy (DAPT), the patients took aspirin 100 mg daily and either clopidogrel 75mg SID (n=35) or ticagrelor 90 mg BID (n=45).
Assessment of Platelet Activity
The assessment of residual platelet reactivity was performed by utilizing VerifyNow assay (“Instrumentation laboratory”, MA, US). For assessment of platelet activity venous blood was used, drained on the 3rd day after the beginning of DAPT therapy in vacuum vials 2 mL with 3.2% sodium citrate. The level of aggregation of platelets is measured in P2Y12 Reactivity Units or percentage of inhibition. The study was conducted within 1 hour after the whole venous blood sample was drained. The therapeutic range of P2Y12-inhibitors when measured with VerifyNow P2Y12 assay are as follows: PRU>208 – the risk of thrombotic events, PRU<95 – bleeding risk, 95<PRU<208 – adequate response.
Genotyping
Blood (2 mL each) for DNA analysis was sampled from the peripheral vein using ethylene diamine tetra acetate (EDTA) tubes (VACUETTE®, Greiner Bio-One, Austria). A panel of 18 SNPs was selected. The following 17 SNPs were selected for clopidogrel:ABCB1 (C3435T, rs1045642), CYP2C19*2 (681G > A, rs4244285), CYP2C19*3 (636G > A, rs4986893), CYP2C19*17 (C-806T, rs1224856), CYP3A4*22 (C>T, rs35599367), CYP3A4 (20239G>A, rs2242480), CYP3A5*3 (A6986G, rs776746), CYP4F2 (C > T, Val433Met, rs2108622), CES1 (А-33С, rs2244613), PON1 (Q192R, rs662), IGTB3 (rs5918), P2Y12 (rs2046934), P2Y12R (A>G, rs3732759), PEAR1 (C>Т, rs41273215), PEAR1 (C>T, rs57731889), B4GALT2 (C>Т, rs1061781), ABCG2 (G>T, rs2231142). The following 6 SNPs were selected for ticagrelor: ABCB1 (C3435T, rs1045642), CYP3A5*3 (A6986G, rs776746), CYP3A4*22 (C>T, rs35599367), CYP3A4 (20239G>A, rs2242480), P2Y12 (rs2046934), SLCO1B1 (T521C, rs4149056). Real-time polymerase chain reaction with commercially available assays («Sintol», Russia; Thermo Fisher Scientific, USA) was performed utilizing Real-Time CFX96 Touch amplifier (Bio-Rad Laboratories, Inc., USA).
MicroRNA Testing
Blood was collected into sterile EDTA tubes and centrifuged for 10 min at 2000g. The obtained supernatant was transferred to sterile 2 mL tubes and stored before use at −80 °C. Total RNA, including miRNA, was extracted using QIAzol lysis reagent and the miRNeasy Serum/Plasma kit (Qiagen, Hilden, Germany) according to the recommended protocol. QIAzol was added to 300 µL of plasma in a volume ratio of 3:1. After chloroform was added to the tube and centrifuged to separate phases, the aqueous phase was transferred to a new tube, and 1.5 times the volume of 100% ethanol was added. The solution containing RNA was loaded into the miRNeasy column and further washed according to the manufacturer’s instructions. The final volume of elution was 20 µL. Concentration and purity of the obtained RNA were measured on the NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, New York, USA). The extraction process was repeated for each sample until a sufficient amount of RNA was obtained for the next steps. Reverse transcription was carried out with the MiScript II RT Kit (Qiagen) according to the manufacturer’s protocol. Expression levels of miR-142, miR-126, miR-223, miR-150, miR-29, miR-34 were estimated with quantitative reverse transcription polymerase-chain reaction (qRT-PCR) using the MiScript SYBR Green PCR Kit (Qiagen) and presynthesizedmiScript Primer Assay (Qiagen) primers. qRT-PCR was performed with three repetitions for the analyzed miRNA in the volume of the reaction mixture of 12 µL on the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, USA) according to the manufacturer’s recommended program (15 min at 95 °C to activate the HotStarTaq DNA Polymerase and 40 three-step cycles (94 °C – 15 s, 55 °C – 30 s, 70 °C – 30 s)). Relative quantifications were calculated using the comparative Ct method (2 ΔΔCt).
Activity of CYP3A4 Isoenzyme
To assess the activity of CYP3A4, patients gave samples of morning urine with a volume of 5 mL, collected in tubes without preservative at the time of the blood draw. Samples were stored frozen at −20 °C. The activity of CYP3A4 was expressed as the ratio of the concentrations of cortisol and 6β-hydroxycortisol, the formation of which occurs under the influence of these isoenzymes. Cortisol and its metabolite were assessed by chromatography–mass spectrometry on an Agilent G1978B Multimode Source high performance liquid chromatograph for 6410 Triple Quad LC/MS (Agilent Technologies, Inc., USA).
Statistics
Data analysis was carried out in the statistical package IBM SPSS Statistics 20.0. All quantitative variables were tested for normal distribution by the Shapiro–Wilk criterion, resulting in abnormal data distribution, except for miR-34, miR-223 expression levels and Base PRU. For the subsequent analysis of quantitative variables between subgroups, nonparametric criteria (Mann-Whitney, Kruskal-Wallis) were applied. Independent Samples t-test and one-way ANOVA were utilized for miR-34, miR-223 and Base PRU variables. A p-value < 0.05 was considered significant. In order to establish the connection between micro-RNA expression levels and platelet reactivity, linear regression was carried out. For the regression analysis, the following variables were selected as the dependent variables: miR-142, miR-126, miR-223, miR-150, miR-29, miR-34. There were no differences from Hardy-Weinberg equilibrium (Table 1).
 |
Table 1 Genotyping Data and Connected miRNA Expression Levels |
Results
MicroRNAs and Genes
The polymorphisms of the P2Y12-inhibitors ADME genes that demonstrated statistically significant connection with miRNA expression levels are as follows (see Table 1): P2Y12R (A>G, rs3732759) and miR-29 (p=0.017), miR-34 (p=0.003); CYP2C19*17 (C-806T, rs1224856) and miR-142 (p=0.012); PON1 (Q192R, rs662) and miR-29 (p=0.004), ABCG2 (G>T, rs2231142) and miR-34 (p=0.007). Therefore, miR-29, miR-34, miR-142 can be used as biomarkers of altered function polymorphisms (see Figure 1).
 |
Figure 1 Connections between miRNAs and polymorphisms of the ADME genes for P2Y12-inhibitors. |
MicroRNAs and Platelet Reactivity Assessment
MiRNAs expression levels showed connection with the results of the platelet reactivity assessment utilizing VerifyNow assay (“Instrumentation laboratory”, MA, US) (see Table 2). MiR-126 (β coefficient=−0.076, SE=0.032, p=0.021), miR-223 (β coefficient=−0.089, SE=0.041, p=0.032), miR-29 (β coefficient=−0.042, SE=0.018, p=0.026), miR-142 (β coefficient=−0.072, SE=0.026, p=0.008) have the potential to be used as biomarkers and to substitute platelet reactivity testing.
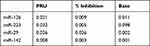 |
Table 2 P-values of the Linear Regression Analysis Between miRNA Expression Levels and Results of the Platelet Reactivity Testing |
Discussion
In this article, six miRNAs and eighteen gene polymorphisms and their connection to platelet reactivity have been assessed. There have been few publications which described the possible connection between miRNA expression levels and resistance to P2Y12-inhibitors.24,28–33 On the basis of these publications six prospective miRNAs have been selected. These are miR-29, miR-34, miR-126, miR-142, miR-150 and miR-223. The aforementioned miRNAs are also noted in the following databases: miRanda,34 PITA,35 miRTarBase,36 Pharmaco-miR,37 TargetscanHuman 7.2.26 These miRNAs are also preselected in the commercially available assays.38 All these facts supported the decision to test the connection between these miRNAs and platelet reactivity as well as loss-of-function genetic polymorphisms.
Recently published trials which assessed the genotype-based approach to prescription of a more potent P2Y12- inhibitor yielded interesting results.16,17 The TAILOR-PCI study demonstrated a 34% reduction in the primary endpoint - a composite of cardiovascular death, MI, stroke, definite or probable stent thrombosis, and severe recurrent ischemia - for patients with genotype-guided approach (4.0% vs 5.9%; adjusted HR 0.66; 95% CI 0.43–1.02). In a post hoc analysis, the researchers found a significant absolute 2.1% benefit in using the genotype-guided strategy at 3 months (HR 0.21; P = 0.001).16 While the TAILOR-PCI study was designed to show superiority of genotyping, another study POPular Genetics compared a genotyping strategy to no-genotyping strategy. It showed non-inferiority of genetic testing to standard care with respect to the primary composite endpoint of all-cause death, MI, definite stent thrombosis, stroke, and PLATO major bleeding (5.1% vs 5.9%; P for non-inferiority < 0.001). Moreover, genetic testing was superior to standard care for the endpoint of PLATO major and minor bleeding (9.8% vs 12.5%; P = 0.04).17
The high number of patients in the study, the number of polymorphisms and the well-established method of P2Y12-inhibitors effectiveness assessment enable miR-29, miR-34, miR-126, miR-142, miR-223 to be used as either biomarkers of platelet reactivity or loss-of-function genetic polymorphisms. Therefore, they can be used as biomarkers of the resistance to P2Y12-inhibitors.
Currently, the genotype-based strategy in prescription of P2Y12-inhibitors lowers the risk of high on-platelet reactivity and MACE.39 Currently the genotype-based strategy in the prescription of P2Y12-inhibitors is only recommended in patients at high risk of thrombotic events and not suitable for all comers as the trials have shown the conflicting results.40 There is an unmet need for a novel complex biomarker that can combine genetic profile and platelet laboratory parameters. MiR-29, miR-34, miR-126, miR-142, miR-223 are such biomarkers.
Limitations
The current study features 6 miRNAs and 80 patients. Although the number of miRNAs is large, the cohort is rather small for pharmacogenomics studies which may, in turn affect statistics and the possibility of false negatives.
Conclusion
This study has revealed new biomarkers of P2Y12- inhibitors resistance: miR-29, miR-34, miR-126, miR-142, miR-223. Vividly demonstrating a connection to on-treatment platelet reactivity, which in the case of miR-29, miR-34, miR-142 is also supported by pharmacogenomics, these novel biomarkers have a potential to be used for P2Y12-inhibitors resistance testing as part of future smart diagnostic tools.
Ethical Approval
All the patients were informed about the purposes of the study and consequently have signed their “consent of the patient”. All investigations conformed to the principles outlined in the Declaration of Helsinki and were performed with permission by the responsible Ethics Committee of Russian Medical Academy of Continuous Professional Education.
Acknowledgments
This study is supported by the state assignment of the Russian Federation “The novel pharmacogenetic biomarkers of major diseases pharmacotherapy” under Project No. 121110800062-6.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Valgimigli M, Bueno H, Byrne RA, et al. 2017 ESC focused update on dual antiplatelet therapy in coronary artery disease developed in collaboration with EACTS. Eur Heart J. 2018;39(3):213–260. doi:10.1093/eurheartj/ehx419
2. Scott SA, Sangkuhl K, Stein CM, et al.; Clinical Pharmacogenetics Implementation Consortium. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94:317–323. doi:10.1038/clpt.2013.105
3. Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi:10.1056/NEJMoa0808227
4. O’Donoghue M, Wiviott SD. Clopidogrel response variability and future therapies: clopidogrel: does one size fit all? Circulation. 2006;114:e600–e606. doi:10.1161/CIRCULATIONAHA.106.643171
5. So DY, Wells GA, McPherson R, et al. A prospective randomized evaluation of a pharmacogenomic approach to antiplatelet therapy among patients with St-elevation myocardial infarction: the RAPID STEMI study. Pharmacogenomics J. 2016;16:71–78. doi:10.1038/tpj.2015.17
6. Notarangelo FM, Maglietta G, Bevilacqua P, et al. Pharmacogenomic approach to selecting antiplatelet therapy in acute coronary syndromes: PHARMCLO trial. J Am Coll Cardiol. 2018;71(17):1869–1877. 7. doi:10.1016/j.jacc.2018.02.029
7. Cavallari LH, Franchi F, Rollini F, et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med. 2018;16(1):92. doi:10.1186/s12967-018-1469-8
8. Klein MD, Lee CR, Stouffer GA. Clinical outcomes of CYP2C19 genotype-guided antiplatelet therapy: existing evidence and future directions. Pharmacogenomics. 2018;19:1039–1046. doi:10.2217/pgs-2018-0072
9. Wallentin L, Becker RC, Budaj A, et al.; PLATO Investigators. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 361;2009:1045–1057. doi:10.1056/NEJMoa0904327
10. Wiviott SD, Braunwald E, McCabe CH, et al.; TRITONTIMI 38 Investigators. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357:2001–2015.9. doi:10.1056/NEJMoa0706482
11. January CT, Wann LS, Calkins H, et al. 2019 AHA/ACC/HRS focused update of the 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. J Am Coll Cardiol. 2019;74(1):104–132. doi:10.1016/j.jacc.2019.01.011
12. Mirzaev KB, Rytkin E, Grishina EA, et al. The impact of CYP2C19, ABCB1 genes polymorphisms and CYP3A4 isoenzyme activity on the incidence of stent implantation complications for patients with an acute coronary syndrome. JACC. 2017;10(3 Supplement):S6.
13. Collet JP, Hulot JS, Anzaha G, et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover Clovis-2 (Clopidogrel and Response Variability Investigation Study 2). JACC. 2011;4(4):392–402. doi:10.1016/j.jcin.2011.03.002
14. Mega JL, Hochholzer W, Frelinger AL, et al. Dosing clopidogrel based on CYP2C19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA. 2011;306(20):2221–2228. doi:10.1001/jama.2011.1703
15. Aradi D, Storey RF, Komocsi A, et al. Expert position paper on the role of platelet function testing in patients undergoing percutaneous coronary intervention. Eur Heart J. 2014;35(4):209–215. doi:10.1093/eurheartj/eht375
16. Pereira NL, Rihal CS, So DYF, et al. Clopidogrel pharmacogenetics state-of-the-art review and the TAILOR-PCI study. Circ Cardiovasc Interv. 2019;12:4. doi:10.1161/CIRCINTERVENTIONS.119.007811
17. Claassens DMF, Vos GJA, Bergmeijer TO, et al. A genotype-guided strategy for oral P2Y12 inhibitors in primary PCI. N Engl J Med. 2019;381(17):1621–1631. doi:10.1056/NEJMoa1907096
18. Tantry US, Bonello L, Aradi D, et al. Consensus and update on the definition of on-treatment platelet reactivity to adenosine diphosphate associated with ischemia and bleeding. J Am Coll Cardiol. 2013;62(24):2261–2273. doi:10.1016/j.jacc.2013.07.101
19. Swen JJ, Nijenhuis M, De Boer A, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673. doi:10.1038/clpt.2011.34
20. Roberts JD, Wells GA, Le May MR, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet (London, England). 2012;379(9827):1705–1711. doi:10.1016/S0140-6736(12)60161-5
21. Rytkin E, Mirzaev KB, Bure IV, Sychev DA. Selection of miRNAs for clopidogrel resistance prediction. Meta Gene. 2020;25. doi:10.1016/j.mgene.2020.100745
22. Chen YC, Lin FY, Lin YW, et al. Platelet microRNA 365-3p expression correlates with high on-treatment platelet reactivity in coronary artery disease patients. Cardiovasc Drugs Ther. 2019;33(2):129–137. doi:10.1007/s10557-019-06855-3
23. Zhou WL, Mo ZZ, Xiao FY, et al. microRNA-605 rs2043556 polymorphisms affect clopidogrel therapy through modulation of CYP2B6 and P2RY12 in acute coronary syndrome patients. Platelets. 2019;30:1–9. doi:10.1080/09537104.2019.1549810
24. Chyrchel B, Totoń-żurańska J, Kruszelnicka O, et al. Association of plasma miR-223 and platelet reactivity in patients with coronary artery disease on dual antiplatelet therapy: a preliminary report. Platelets. 2015;26(6):593–597. doi:10.3109/09537104.2014.974527
25. Parker WAE, Schulte C, Barwari T, et al. Aspirin, clopidogrel and prasugrel monotherapy in patients with type 2 diabetes mellitus: a double-blind randomised controlled trial of the effects on thrombotic markers and microRNA levels. Cardiovasc Diabetol. 2020;19(1):3. doi:10.1186/s12933-019-0981-3
26. Agarwal V, Bell GW, Nam J-W, et al. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4(4):e05005. doi:10.7554/eLife.05005
27. Rytkin EI, Mirzaev KB, Bure IV, et al. Micro-RNA as a new biomarker of activity of the cytochrome system P-450: significance for predicting the antiplatelet action of P2Y12 receptor inhibitors. Ter Arkh. 2019;91(8):115–117.
28. Carino A, De Rosa S, Sorrentino S, et al. Modulation of circulating microRNAs levels during the switch from clopidogrel to ticagrelor. Biomed Res Int. 2016;2016:3968206. doi:10.1155/2016/3968206
29. Tian HS, Zhou QG, Shao F. Relationship between arterial atheromatous plaque morphology and platelet–associated miR–126 and miR–223 expressions. Asian Pac J Trop Med. 2015;8(4):309–314. doi:10.1016/S1995-7645(14)60336-9
30. Shi R, Zhou X, Ji WJ, et al. The emerging role of miR-223 in platelet reactivity: implications in antiplatelet therapy. Biomed Res Int. 2015;2015:981841. doi:10.1155/2015/981841
31. Kaudewitz D, Skroblin P, Bender LH, et al. Association of microRNAs and YRNAs with platelet function. Circ Res. 2016;118(3):420–432. doi:10.1161/CIRCRESAHA.114.305663
32. Ambrose AR, Alsahli MA, Kurmani SA, Goodall AH. Comparison of the release of microRNAs and extracellular vesicles from platelets in response to different agonists. Platelets. 2018;29(5):446–454. doi:10.1080/09537104.2017.1332366
33. Stakos DA, Gatsiou A, Stamatelopoulos K, Tselepis AD, Stellos K. Platelet microRNAs: from platelet biology to possible disease biomarkers and therapeutic targets. Platelets. 2013;24(8):579–589. doi:10.3109/09537104.2012.724483
34. Betel D, Koppal A, Agius P, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi:10.1186/gb-2010-11-8-r90
35. Kertesz M, Iovino N, Unnerstall U, et al. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi:10.1038/ng2135
36. Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):
37. Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N. Pharmaco-miR: linking microRNAs and drug effects. Brief Bioinform. 2014;15(4):
38. Krammer TL, Mayr M, Hackl M. microRNAs as Promising Biomarkers of Platelet Activity in Antiplatelet Therapy Monitoring. International Journal of Molecular Sciences. 2020;21(10):3477. doi:10.3390/ijms21103477
39. Gower MN, Ratner LR, Williams AK, Rossi JS, Stouffer GA, Lee CR. Clinical utility of CYP2C19 genotype-guided antiplatelet therapy in patients at risk of adverse cardiovascular and cerebrovascular events: a review of emerging evidence. Pharmgenomics Pers Med. 2020;13:239–252. doi:10.2147/PGPM.S231475
40. Akhtar T, Bandyopadhyay D, Ghosh RK, Aronow WS, Lavie CJ, Yadav N. Advances in the pharmacogenomics of antiplatelet therapy. Am J Ther. 2020;27(5):e477–e484. doi:10.1097/MJT.0000000000001013
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
