Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNA Derived from Circulating Exosomes as Noninvasive Biomarkers for Diagnosing Renal Cell Carcinoma
Authors Xiao CT , Lai WJ, Zhu WA, Wang H
Received 16 July 2020
Accepted for publication 2 October 2020
Published 23 October 2020 Volume 2020:13 Pages 10765—10774
DOI https://doi.org/10.2147/OTT.S271606
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Chu-Tian Xiao,* Wen-Jie Lai,* Wei-An Zhu, Hua Wang
Department of Urology, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, People’s Republic of China, 510630
*These authors contributed equally to this work
Correspondence: Hua Wang
Department of Urology, The Third Affiliated Hospital of Sun Yat-sen University, Guangzhou 510630, People’s Republic of China
Tel +86-20-85253333
Fax +86-20-85253336
Email [email protected]
Purpose: Renal cell carcinoma (RCC) is the most common type of kidney cancer in adults. Exosomes are membrane-enclosed extracellular vesicles, and exosomal RNA can be a biomarker for cancer diagnosis and prognosis in RCC patients. We aim to identify differences in miRNA expression profiles in peripheral blood exosomes between RCC patients and healthy subjects as well as to investigate novel markers of RCC.
Methods: We performed exosomal miRNA sequencing of plasma samples obtained from five RCC patients and five control subjects, subsequently 22 RCC patients and 16 control subjects were investigated using qPCR to confirm the differential miRNA which from plasma exosomal RNA sequencing. ROC curves were constructed to assess the diagnostic accuracy of exosomal miRNAs as diagnostic biomarkers of RCC.
Results: Exosomes were isolated with the exoeasy maxi kit and confirmed using TEM and NTA. They have a spherical structure with a diameter of approximately 40– 180 nm. The exosomal miRNA sequence results showed that a total of 2357 miRNAs were detected, and 245 miRNAs were differentially expressed between RCC patients and healthy controls (p< 0.001, average counts > 5, log|fc|> 1). Further analysis revealing that, versus the control, 17 miRNAs are up-regulated and 5 miRNAs are down-regulated under selection conditions with average miRNAs counts > 100. qPCR was performed using 38 subjects—the results showed that the expression levels of hsa-mir-149-3p and hsa-mir-424-3p were upregulated; the expression levels of hsa-mir-92a-1-5p were significantly downregulated in the plasma exosomes of RCC. For diagnosis of RCC, the AUC of hsa-mir-92a-1-5p, hsa-mir-149-3p and hsa-mir-424-3p was 0.8324, 0.7188 and 0.7727, with the sensitivity of 0.875, 0.750 and 0.750, and the specificity of 0.773,0.727 and 0.818, respectively, at the best cutoff value.
Conclusion: Our study revealed that the expression levels of hsa-mir-92a-1-5p, hsa-mir-149-3p and hsa-mir-424-3p were significantly abnormal in RCC patients, which may be novel biomarkers for RCC diagnosis.
Keywords: renal cell carcinoma, exosome, miRNAs
Introduction
Renal cell carcinoma (RCC) originates from renal tubular epithelial cells and is one of the most common malignant tumors in the urinary system.1 Early renal cancer patients have no obvious symptoms, and the diagnosis mainly depends on imaging examinations. A population-based study showed that 21% of the patients had metastatic disease, and 20% of the primary non-metastatic patients had recurrent diseases during a 5-year follow-up period.2 Surgical resection is the only effective current cure for renal cancer.
MicroRNA (miRNA) is completely or partially complementary to the 3ʹ untranslated sequence (3ʹ-UTR) of target messenger RNA resulting in degradation or post-transcriptional translation inhibition of target messenger RNA. This regulates the expression of target gene at the post-transcriptional level.3 Studies have confirmed that there are abundant and stably expressed miRNA in human peripheral blood circulation, and specific circulating miRNA can also be transmitted between different cells of exosomes.4
Exosomes are small vesicles with a diameter of 30–150 nm, which are secreted by a variety of cells and encapsulated in a lipid bilayer membrane.5 Exosomes exist in a variety of body fluids such as peripheral blood, urine, and ascites. Exosomes were once considered to be cellular waste, but subsequent studies have gradually found that exosomes can carry a variety of signaling molecules and bioactive substances that participate in a variety of biological processes such as body immunity, intercellular communication, and tumor occurrence/development.6 In recent years, exosomes have become a research hotspot in the field of urological cancer such as prostate cancer and bladder cancer. Studies have reported abnormal expression of exosomal miR-210, miR-126-3p, miR-449a, miR-34b-5p, and miR-1233 in RCC patients.7–9 Thus, exosomal miRNAs might be a diagnostic biomarker in RCC patients.
Here, exosome miRNA expression profiles of peripheral blood samples between RCC patients and healthy subjects were tested via high-throughput sequencing to determine whether exosomal miRNAs can be used as markers for RCC diagnosis.
Materials and Methods
Patients and Samples
RCC patients and controls were recruited from The Third Affiliated Hospital of Sun Yat-sen University. In the first stage, ten samples were used for exosomal RNA sequencing, which included five RCC patients and five control subjects. Disease information of RCC patients is shown in Table 1, and the control subjects came from healthy patients without tumors and chronic diseases. In the second stage, 38 subjects (16 healthy individuals and 22 RCC) were investigated using qPCR to confirm the differential miRNA from plasma exosomal RNAs prior to receiving drug treatment. Patients were required to provide medical history and receive laboratory tests and to have a clear histological pathological diagnosis, and the details are shown in Table 2. The registrants in the control group excluded tumors, inflammation, severe heart failure and moderate and severe anemia, but some registrants in the control individuals were complicated with diabetes (n=2), hypertension (n=3) and coronary heart disease (n=3). Blood was collected from the elbow vein with an EDTA-K2 tube, for about 4–5mL a sample. Each sample was centrifuged at 3000 rpm for 10 min at 4°C to remove cellular debris, and the isolated plasma samples were stored at −80°C until RNA isolation. The study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China. All patients and control subjects enrolled in the study provided informed consent, in accordance with the Declaration of Helsinki.
 |
Table 1 The Clinical Characteristics of RCC Patients Who Received Exocrine RNA Sequencing |
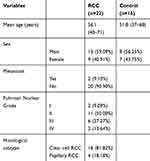 |
Table 2 The Clinical Characteristics of Patients Who Receive Exocrine PCR Test |
Exosomal RNA Extraction and Sequencing Library Preparation
Exosomes were isolated with the exoEasy Maxi Kit (QIAGEN, Catalog Number: 76064, Germantown, MD, USA) according to the manufacturer’s instructions. Total RNA was isolated from exoEasy kit columns following the protocol of the exoRNeasy SerumKit (QIAGEN, Catalog Number: 77064, Germantown, MD, USA). The RNA yield and size range were analyzed on an Agilent 2100 Bioanalyzer. Extracted RNA was used to prepare miRNA-focused NGS libraries with QIAseq miRNA Library Kit (QIAGEN, Catalog Number: 331505, Germantown, MD, USA). The sequencing was performed on an Illumina NovaSeq 6000 System at the HaploX Biotechnology Co., LTD (Shenzhen, China). The sequencing data was analyzed with the QIAseq miRNA quantification platform using unique molecular index counts.
ZetaView Nanoparticle Tracking Analysis (NTA)
Sample exosomes were analyzed for size and concentration using the ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany). The exosomal sample was diluted in PBS to obtain a measurable concentration between 1.0x108 and 1.0x109 particles/mL. For analysis, a monochromatic laser beam was applied to the diluted plasma exosomes. Software (ZetaView 8.02.28) analyzed the sample at a constant temperature to determine a final estimate of the size and concentration of the exosomes. The NTA settings were pre-optimized and remained constant between samples. The temperature was held between 20°C and 30°C.
Quantitative Reverse Transcription Polymerase Chain Reaction
Total RNA was extracted from exosomes using a miRNeasy kit (QIAGEN, Germantown, MD, USA) according to the manufacturer’s instructions. The concentration of microRNAs was determined before polymerase chain reaction. In the 20-μL reverse transcription reaction system, we added 2×microRNA RT solution (10 μL), microRNA RT Enzyme mix (2 μL) and microRNA (100 ng). In the 20-μL PCR system, there were 2 ×microRNA qPCR master mix (10 μL), template DNA (2 μL), and the corresponding upstream and downstream primers were 0.5 μL. Reaction conditions: 95°C for 30 s; 95°C for 5 s; 60oC for 30 s; and 60oC for 30 s. This was repeated 40 rounds. Data were calculated and counted using a 2−ΔCt value calculation and miR-16-5p for normalization. The primers for amplification are shown in Table 3
 |
Table 3 The Primers for Amplification |
Sequencing Data Analysis and Statistical Analysis
Sequencing data were processed via a routine bioinformatics analysis method. FastQC package (version 0.11.3) was used to assess sequence length distribution and quality. Adaptor sequences were trimmed using Btrim, and all reads without adaptors or shorter than 16 bases were discarded. Reads were aligned in the recent version of miRBase, and mapping was performed using Bowtie. Principal component analysis (PCA) application for RNA-seq was used for exosome classification. Differential gene expression (DGE) analysis was performed with the Bioconductor Package DESeq2 (version 1.8.1). Hierarchical clustering, PCA, and visualization of significantly regulated miRNAs were carried out in the R (version 3.5.1) statistical language and R Studio software. Receiver operating characteristic (ROC) curves were constructed and the area under the ROC curve (AUC) was computed to assess the diagnostic power of plasma exosomal miRNAs. P<0.05 was indicated a statistically significant difference.
Results
Characterization of Isolated Exosomes
The successful extraction of exosomes is an important prerequisite for analyzing the characteristics of exosomal miRNA expression. Therefore, we used NTA to determine the size and concentration of exosomes. As shown (Figure 1), the diameters of plasma exosomes were 50 nm to 200 nm. The concentration of exosomes was 7.2 × 1010cells/mL. (This figure shows the result of 1000-fold dilution).
Exosomal miRNA Profiling
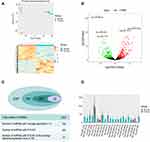
We performed plasma exosomal miRNA profiling for five patients with RCC and five matched healthy controls. Each microRNA expression level (counts) was determined. The integrated Unique Molecular Indices (UMIs), which is packed in the QIAseq miRNA Library Kit, enabled quantification of individual miRNA molecules. The UMIs tag each miRNA during the miRNA library construction stage eliminating PCR and sequencing bias. The integrated data analysis is available through GeneGlobe for miRNA mapping, data analysis, and differential expression analysis. PCA analysis was performed, and the presented variance was analyzed by PC1 and PC2 at 45.03% and 11.57%, respectively. These accounted for a total of 56.6% of the total variance (Figure 2A).
The content and distribution of microRNA in exosomes differs greatly from that in cells. A total of 2357 miRNAs were detected. We removed the low average expression of microRNAs, and 740 miRNAs average counts >5. Differential expression of miRNAs was performed using 740 miRNAs, and the results revealed that 245 miRNAs were differentially expressed between RCC patients and healthy controls (P<0.001, log|FC|>1) with 140 miRNAs being up-regulated and 105 miRNAs down-regulated in RCC (Figure 2C). A volcano plot shows up-regulated and down-regulated miRNAs (Figure 2B) Considering the abundance of miRNAs in the exosomes, setting the selection condition to have average counts that are >100 caused 22 miRNAs to be filtered out with 17 miRNAs up-regulated and five miRNAs down-regulated (Figure 2D).
GO Enrichment Annotation Analysis and KEGG Enrichment Pathway Analysis
After screening for the miRNAs, many of them were found to have functional genes closely related to RCC, and the miRNAs that regulate these functional genes also were found to contain the differentially expressed miRNAs previously found. Target genes corresponding to differentially expressed miRNAs (P<0.001, log|FC|>1) were predicted by starBase, and we performed GO enrichment analysis on all differentially expressed miRNA using DAVID. The target miRNAs have a wide range of biological functions. GO enrichment results showed that upregulated miRNAs were the most significant enrichment term in GO:0003713, GO:0001077, GO:0001228, GO:0004674, GO:0000982, GO:0005667, and GO:0031252 (Figure 3A). The downregulated miRNAs were the most significant enrichment term in GO:0001228, GO:0004842, GO:0019787, GO:0004674, GO:0000982, GO:0005667 (Figure 3B). The KEGG annotation analysis for upregulated miRNAs in RCC indicated hsa05215 in prostate cancer, hsa05131 in Shigellosis, hsa04550 as a signaling pathway regulating pluripotency of stem cells, hsa05100 for bacterial invasion of epithelial cells, hsa04360 in axon guidance, and hsa04390 in hippo signaling pathways were involved (Figure 3C). The KEGG pathway analysis revealed that those downregulated miRNAs were significantly enriched in multiple pathways involved in hsa04550, hsa05211 in renal cell carcinoma, hsa05205 for proteoglycans in cancer, hsa04722 in neurotrophin signaling pathways, hsa04390, as well as hsa04350 for TGF-beta signaling pathways (Figure 3D).
Validation of Differentially Expressed Plasma Exosomal miRNAs by qPCR Analysis
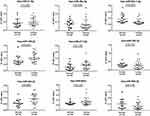
We selected miRNAs with less than 1000 numbers from the differential genes for PCR verification, and qPCR was performed in 38 subjects (16 healthy individuals and 22 RCC patients). Nine differentially expressed miRNAs were analyzed by qPCR, and five had significant differences (P<0.05) versus those in healthy individuals. The expression levels of hsa-miR-149-3p and hsa-miR-424-3p were found to be upregulated, and the expression levels of hsa-miR-92a-1-5p were significantly downregulated in the plasma exosomes of RCC. The expression levels of hsa-miR-21-5p, hsa-miR-29a-3p, hsa-miR-342-3p, and hsa-miR-885-3p were not significantly changed (Figure 4).
The Diagnostic Value of Plasma Exosomal miRNAs for RCC
The area under the ROC curve can evaluate the accuracy of a diagnostic method. In this study, ROC curves were constructed to assess the diagnostic accuracy of plasma exosomal miRNAs as plasma tumor markers. We performed ROC curve analysis on nine different miRNAs. The results showed that the AUCs of the 9 miRNAs ranged from 0.6264 to 0.8324. The AUC of hsa-miR-92a-1-5p was the highest at 0.8324. At the optimal cutoff value, hsa-miR-92a-1-5p offered 87.5% specificity and 77.3% sensitivity; its diagnostic accuracy is better than other miRNAs. The hsa-miR-424-3p also offered a high AUC value of 0.7727 with 75.0% specificity and 81.8% sensitivity. Detailed ROC results are shown in Figure 5.
 |
Figure 5 ROC analysis for distinguishing RCC cases from controls using plasma exosomal miRNAs. |
Discussion
Exosomes are sub-vesicle structures secreted by various cells in the body that carry a variety of protein and nucleic acid substances derived from cells. These structures can be easily obtained from peripheral blood and other body fluids and are used for tumor diagnoses.10 Many studies have reported that miRNAs are contained in exosomes involved in the occurrence of cancer. They can be used as a biomarker for cancer diagnosis. Fabbri et al11 found that miRNAs in lung cancer-derived exosomes promote tumor cell growth and metastasis by triggering Toll-like receptors. Zhou et al12 found that exosomes mediate the secretion of miR-105 from tumor cells that effectively disrupt the tight junction and integrity of the natural barrier in the vascular endothelial cell layer; these natural barriers can counter the metastasis of tumor cells. Another study found that the expression level of plasma exosome miR-141 is increased in prostate cancer patients and is expected to be a molecular diagnostic marker for prostate cancer.13 Of course, studies on exosomes and RCC have also been reported, involving mechanistic research and disease diagnosis. Lindoso et al14 found that exosomes derived from a RCC not only affect cells in the tumor microenvironment but also cells distant cells. Diao et al15 reported that RCC-derived exosomes are rich in Hsp70 and that Hsp70 promotes the immunosuppressive activity of myeloid-derived cells against the body by inducing the release of pro-inflammatory cytokines and tumor growth factors—this promotes tumorigenesis. This insight provides a new target for future research in cancer therapy.
In this study, we determined differentially expressed exosomal miRNAs between the RCC and control groups, and the distribution of these differentially expressed exosomal miRNAs was visualized in volcano plots. We excluded miRNAs with low expression and identified 22 differential miRNAs. We subsequently performed qRT-PCR verification on 9 miRNAs including hsa-miR-149-3p, hsa-miR-424-3p, hsa-miR-92a-1-5p, hsa-miR-21-5p, hsa-miR-29a-3p, hsa-miR-211-5p, hsa-miR-342-3p, hsa-miR-663a, and hsa-miR-885-3p. The results revealed that exosomal hsa-miR-149-3p and hsa-miR-424-3p were upregulated, and the expression levels of exosomal hsa-miR-92a-1-5p were significantly downregulated in the plasma exosomes of RCC. The ROC analysis showed that the AUC of hsa-miR-92a-1-5p was 0.8324 with 87.5% specificity and 77.3% sensitivity at the optimal cutoff value; the hsa-miR-424-3p yielded a high AUC value of 0.7727 with 75.0% specificity and 81.8% sensitivity. Other miRNAs do not have satisfactory sensitivity and specificity with AUC values of 0.6264 to 0.7188. For example, miR-92a is one of the most studied miR-17-92a gene clusters and plays an important role in modulating physiological processes. Some studies have shown that miR-92a can promote cell proliferation by negatively regulating the p53 subtype, and it can also regulate apoptosis. Obviously, miR-92a may have the characteristics of oncogenes—a study found that the proliferation of hepatocellular carcinoma-derived cell lines was enhanced by miR-92a and inhibited by the anti-miR-92a antagomir.16 Experimental studies also found that miR-92a can promote the cell activity and proliferation of liver cancer cells, distant metastasis, and tumor development.17,18 MiR-92a can also reduce the expression of E-cadherin and increase the expression of vimentin indicating that it can promote the development of EMT process.19 However, there are few reports on the study of exosomal miR-92a. In this project, miRNA sequencing and qPCR showed that the miR-92a derived from peripheral blood exosomes in patients with RCC was lower than that in the control group. It could be used for the diagnosis of RCC. We speculate that the content and type of miRNA in exosomes must be different from their mother cells, and there are significant differences in exosomal miRNAs in different tumors. The miR-424 has important effects on cell differentiation, proliferation, cycle, migration, and angiogenesis, and the above-mentioned disorders of cell function are often an important part of tumorigenesis and malignant process.20 Studies have shown that there is abnormal expression of miR-424 in a variety of tumor tissues, and abnormal expression in chronic lymphocytic leukemia,21 breast cancer,22 ovarian cancer,23 and cervical cancer.24 This study found that exosomal hsa-miR-424 has diagnostic value in RCC suggesting that exosomal hsa-miR-424 has a direct or indirect relationship with the occurrence and development of RCC exosomal hsa-miR-424.
However, our research has several limitations. The molecular mechanism of different histological RCC subtype may be different, and so the circulating exosomes may be different. In this study, a small number of cases were included in the screening stage, with 4 Clear cell RCC and 1papillary RCC. In the validation stage, most cases were Clear cell RCC, which could not fully reflect the real situation of RCC. If the study focused on the subtype of clear cell RCC at the initial stage of design and included more cases, the conclusion will be more convincing. In addition, this study did not further analyze exosomal miRNAs and clinical information, which is one of the shortcomings of this study. In addition, the inclusion of the control group should also be more rigorous, in order to make the conclusion more solid.
In conclusion, this exosome sequencing project found abnormal expression of hsa-miR-149-3p, hsa-miR-211-5p, hsa-miR-424-3p, hsa-miR-663a, and hsa-mi-92a-1-5p in RCC. The ROC curve analysis revealed that plasma exosome hsa-mi-92a-1-5p, hsa-miR-149-3p, and hsa-miR-424-3p could be used as promising biomarkers for detecting RCC. However, further validation using a larger cohort is needed to confirm the results of our study.
Ethics
The study was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University, Guangzhou, China. All patients and control subjects enrolled in the study provided informed consent, in accordance with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was approved by the Youth Program of National Natural Science Foundation of China (Grant number: 81902617).
Disclosure
The authors declare that they have no competing interests.
References
1. Muglia VF, Prando A. Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol Bras. 2015;48(3):166–174. doi:10.1590/0100-3984.2013.1927
2. Dabestani S, Thorstenson A, Lindblad P, et al. Renal cell carcinoma recurrences and metastases in primary non-metastatic patients: a population-based study. World J Urol. 2016;34(8):1081–1086. doi:10.1007/s00345-016-1773-y
3. Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi:10.1016/j.cell.2007.12.024
4. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–6333. doi:10.1073/pnas.0914843107
5. Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;9(59):e3037.
6. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi:10.1016/j.tcb.2015.01.004
7. Butz H, Nofech-Mozes R, Ding Q, et al. Exosomal microRNAs are diagnostic biomarkers and can mediate cell-cell communication in renal cell carcinoma. Eur Urol Focus. 2016;2(2):210–218. doi:10.1016/j.euf.2015.11.006
8. Dias F, Teixeira AL, Nogueira I, et al. Extracellular vesicles enriched in hsa-miR-301a-3p and hsa-miR-1293 dynamics in clear cell renal cell carcinoma patients: potential biomarkers of metastatic disease. Cancers. 2020;12(6):1450. doi:10.3390/cancers12061450
9. Dias F, Teixeira AL, Nogueira I, et al. Plasma extracellular vesicle-derived TIMP-1 mRNA as a prognostic biomarker in clear cell renal cell carcinoma: a pilot study. Int J Mol Sci. 2020;21(13):4624. doi:10.3390/ijms21134624
10. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138
11. Fabbri M. TLRs as miRNA receptors. Cancer Res. 2012;72(24):6333–6337. doi:10.1158/0008-5472.CAN-12-3229
12. Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–515. doi:10.1016/j.ccr.2014.03.007
13. Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther. 2016;9:139–148.
14. Lindoso RS, Collino F, Camussi G. Extracellular vesicles derived from renal cancer stem cells induce a pro-tumorigenic phenotype in mesenchymal stromal cells. Oncotarget. 2015;6(10):7959–7969. doi:10.18632/oncotarget.3503
15. Diao J, Yang X, Song X, et al. Exosomal Hsp70 mediates immunosuppressive activity of the myeloid-derived suppressor cells via phosphorylation of Stat3. Med Oncol. 2015;32(2):453. doi:10.1007/s12032-014-0453-2
16. Shigoka M, Tsuchida A, Matsudo T, et al. Deregulation of miR-92a expression is implicated in hepatocellular carcinoma development. Pathol Int. 2010;60(5):351–357. doi:10.1111/j.1440-1827.2010.02526.x
17. He G, Zhang L, Li Q, et al. miR-92a/DUSP10/JNK signalling axis promotes human pancreatic cancer cells proliferation. Biomed Pharmacother. 2014;68(1):25–30. doi:10.1016/j.biopha.2013.11.004
18. Zhang G, Zhou H, Xiao H, et al. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci. 2014;59(1):98–107. doi:10.1007/s10620-013-2858-8
19. Lu C, Shan Z, Hong J, et al. MicroRNA-92a promotes epithelial-mesenchymal transition through activation of PTEN/PI3K/AKT signaling pathway in non-small cell lung cancer metastasis. Int J Oncol. 2017;51(1):235–244. doi:10.3892/ijo.2017.3999
20. Chamorro-Jorganes A, Araldi E, Penalva LO, et al. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31(11):2595–2606. doi:10.1161/ATVBAHA.111.236521
21. Sun YP, Lu F, Han XY, et al. MiR-424 and miR-27a increase TRAIL sensitivity of acute myeloid leukemia by targeting PLAG1. Oncotarget. 2016;7(18):25276–25290. doi:10.18632/oncotarget.8252
22. Wang J, Wang S, Zhou J, et al. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother. 2018;102:147–152. doi:10.1016/j.biopha.2018.03.018
23. Hua F, Li CH, Chen XG, et al. Long noncoding RNA CCAT2 knockdown suppresses tumorous progression by sponging miR-424 in epithelial ovarian cancer. Oncol Res. 2018;26(2):241–247. doi:10.3727/096504017X14953948675412
24. Xu J, Li Y, Wang F, et al. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–987. doi:10.1038/onc.2012.121
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.