Back to Journals » OncoTargets and Therapy » Volume 13
MicroRNA-939 Directly Targets HDGF to Inhibit the Aggressiveness of Prostate Cancer via Deactivation of the WNT/β-Catenin Pathway
Authors Situ J, Zhang H, Jin Z, Li K, Mao Y, Huang W
Received 16 February 2020
Accepted for publication 8 April 2020
Published 18 May 2020 Volume 2020:13 Pages 4257—4270
DOI https://doi.org/10.2147/OTT.S250101
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
This paper has been retracted.
Jie Situ, 1,* Hao Zhang, 1,* Zi Jin, 2 Ke Li, 1 Yunhua Mao, 1 Wentao Huang 1
1Department of Urology, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, People’s Republic of China; 2Department of Hepatological Surgery, The Third Affiliated Hospital of Sun Yat-Sen University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wentao Huang
Department of Urology, The Third Affiliated Hospital of Sun Yat-Sen University, 600 Tianhe Road, Guangzhou 510000, People’s Republic of China
Email [email protected]
Purpose: MicroRNA-939 (miR-939) has crucial roles in several types of human cancer. However, the expression profile and precise functions of miR-939 in prostate cancer (PCa) are still unclear. This study aimed to determine miR-939 expression in PCa and explore its roles in PCa tumorigenesis.
Methods: miR-939 expression was determined in PCa tissues and cell lines using reverse transcription–quantitative polymerase chain reaction. Cell Counting Kit-8, colony formation, and flow cytometric assays were used to determine the role of miR-939 in PCa cell proliferation and apoptosis in vitro, whereas a tumor xenograft model was generated to evaluate the effect of miR-939 on tumor growth in vivo. Transwell assays were performed to investigate whether miR-939 affects the migration and invasiveness of PCa cells.
Results: miR-939 was found to be downregulated in PCa tissues and cell lines, and this downregulation was significantly correlated with tumor stage and lymphatic metastasis. Patients with PCa exhibiting low miR-939 expression had shorter overall survival than those exhibiting high miR-939 expression. Exogenous miR-939 expression suppressed PCa cell proliferation, colony formation, migration, and invasion in vitro; enhanced apoptosis in vitro; and decreased tumor growth in vivo. Investigation of the underlying molecular mechanisms revealed hepatoma-derived growth factor (HDGF) as a direct target gene of miR-939 in PCa. HDGF was found to be significantly upregulated in PCa tissues, and its expression was inversely correlated with miR-939 expression. HDGF silencing and miR-939 upregulation showed similar effects in PCa. Restored HDGF expression counteracted the tumor-suppressive activity of miR-939 overexpression in PCa cells. Furthermore, ectopic miR-939 expression inhibited the WNT/β-catenin pathway activation in PCa both in vitro and in vivo by downregulating HDGF.
Conclusion: miR-939 functions as a tumor suppressor during PCa tumorigenesis by directly targeting HDGF and deactivating the WNT/β-catenin pathway, suggesting the miR-939/HDGF/WNT/β-catenin pathway as an effective target for PCa therapy.
Keywords: microRNA-939, prostate cancer, hepatoma-derived growth factor, WNT/β-catenin signaling
Introduction
Prostate cancer (PCa) is the third most frequently diagnosed human malignant tumor and the seventh leading cause of cancer-related deaths worldwide.1 The morbidity rate of PCa has increased rapidly in China over the past decade.2 It is also the most common male tumor in Europe and USA, with an morbidity of 214 cases per 1000 men.3–5 Currently, the first-line treatment options for PCa are surgical resection, chemotherapy (including targeted therapy), radiotherapy, and hormone therapy.6 Although great progress has been made in the diagnosis and treatment of PCa in recent years, the long-term clinical outcomes of patients with PCa are still unsatisfactory.7,8 Several risk factors, including age, genetic factors, androgen dependence, sex hormones, environmental factors, and diet, have been implicated in PCa pathogenesis.9,10 The activation of oncogenes and the suppression of tumor-suppressing genes also perform major contributions to PCa oncogenesis.11,12 However, the mechanisms involved in the development and progression of PCa are not well understood. Therefore, additional data complementing the existing knowledge about the molecular mechanisms underlying the initiation and progression of PCa are urgently needed and may be useful for developing new therapies for treating this malignant tumor.
MicroRNAs (miRNAs), a group of noncoding short RNAs of 18–22 nucleotides, can selectively interact with the 3′ untranslated region (3′-UTRs) of their target mRNAs, thus promoting their degradation and inducing translation suppression.13 To date, >2000 miRNA genes have been confirmed in the human genome and are estimated to modulate approximately one-third of all human protein-coding genes.14 All the factors involved in physiological and pathological processes, such as cell survival, development, differentiation, metabolism, and carcinogenesis, are regulated by miRNAs.15,16 A plenty of studies showed that miRNAs perform crucial roles in the control of anti-cancer immune response through the regulation of immune checkpoints.17–19 In terms of cancer, miRNAs play either a tumor-suppressive or tumor-promoting role depending on the characteristics of their target genes, and affect a number of malignant characteristics.20–22 Aberrant miRNA expression has been frequently identified in PCa and demonstrated to be closely associated with prostate carcinogenesis, including PCa progression.23–25 In addition, miRNAs are regarded as perfect candidates for identifying tumor diagnosis and prognosis biomarkers.26,27 Therefore, an in-depth exploration of PCa-related miRNAs may offer novel insights into the molecular pathways underlying PCa progression and facilitate the identification of novel targets for anticancer therapy.
At present, serum prostate specific antigen (PSA) is the most commonly used diagnosis biomarker for PCa, yet it still has limitation such as low specificity.21,28 Accordingly, the interest in miRNA research is due to the fact that PSA shows several limitations and an early diagnosis especially in the high risk is needed. The crucial roles of miR-939 have been demonstrated in several types of human cancer, including colorectal,29 gastric,30 and ovarian cancer.31 However, the expression profile and precise roles of miR-939 in PCa remain unclear. Hence, this study investigated miR-939 expression in PCa tissues and cell lines and explored the effect of miR-939 on the malignant phenotype of PCa cells in vitro and in vivo. Moreover, the mechanisms underlying miR-939’s action on PCa progression were explored at the molecular level. Our results may offer opportunities for identifying effective diagnosis biomarker and promising therapeutic strategies to inhibit PCa progression.
Materials and Methods
Patients and Tissue Samples
A total of 58 pairs of PCa tissue samples and their corresponding adjacent normal tissues were collected from patients who received surgical treatment at The Third Affiliated Hospital of Sun Yat-Sen University. None of the patients had been treated with chemotherapy, radiotherapy, or other anticancer therapies before surgery. The status of lymphatic metastasis was determined by the pathological examination. All tissue specimens were stored in liquid nitrogen for subsequent total RNA or protein isolation. All experimental steps involving the use of clinical tissues were approved by the Ethics Committee of the Tongde Hospital of Zhejiang Province and performed in accordance with the Declaration of Helsinki. In addition, written informed consent was obtained from all participating patients.
Cell Culture
Human PCa cell lines (DU145, 22RV1, LNCaP, and PC3) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The culture medium Eagle’s Minimum Essential Medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used for DU145 cells. Cell lines 22RV1and LNCaP were kept in RPMI-1640 Medium (Gibco; Thermo Fisher Scientific, Inc.,), while Ham’s F-12 medium (Gibco; Thermo Fisher Scientific, Inc.,) was used for PC3 cells. All above basal medium was supplemented with 10% fetal bovine serum (FBS) and 1% of penicillin/streptomycin (all from Gibco; Thermo Fisher Scientific,).
A normal prostatic epithelial cell line (RWPE-1; Chinese Academy of Sciences) was cultured in Keratinocyte Serum Free Medium (Gibco; Thermo Fisher Scientific,) containing 10% FBS. All cell lines were cultured at 37°C in a humidified atmosphere comprising 5% CO2.
Oligonucleotides, Plasmids, and Cell Transfection
miR-939 agomir (agomir-939), negative control (NC) agomir (agomir-NC), the hepatoma-derived growth factor (HDGF)-overexpressing plasmid pcDNA3.1-HDGF lacking its 3′-UTR (pc-HDGF), and an empty pcDNA3.1 vector were chemically synthesized by GenePharma (Shanghai, China). Small interfering RNA (siRNA) inhibiting the expression of HDGF (si-HDGF) and NC siRNA (si-NC) were acquired from RiboBio Co., Ltd. (Guangzhou, China). For transfection, the cells were seeded in 6-well plates. The aforementioned nucleic acids were transfected into the cells using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol.
Reverse Transcription–Quantitative Polymerase Chain Reaction (RT–qPCR)
Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen GmbH, Hilden, Germany) and then reverse transcribed using the miScript Reverse Transcription Kit (Qiagen GmbH). After complementary DNA (cDNA) was produced, qPCR was conducted to determine miR-939 expression using the miScript SYBR Green PCR Kit (Qiagen GmbH). To quantify HDGF mRNA expression, cDNA was synthesized from total RNA with the PrimeScript RT Reagent Kit (Takara Biotechnology Co., Ltd., Dalian, China). This cDNA was then subjected to qPCR with SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). Small nuclear RNA U6 and GAPDH served as the internal references for miR-939 and HDGF mRNA, respectively. The relative gene expression was analyzed using the 2−∆∆Cq method.
Cell Counting Kit-8 (CCK-8) and Colony Formation Assays
Transfected cells were harvested at 24 h posttransfection and used for evaluating cell proliferation. For the CCK-8 assay, cells were seeded in 96-well plates at a density of 2 × 103 cells per well. At 0, 24, 48, and 72 h after seeding, the CCK-8 assay was conducted by adding 10 μL of the CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) into each well. Following 2 h of incubation at 37°C, the absorbance was measured at 490 nm using a SUNRISE Microplate Reader (Tecan Group, Ltd., Mannedorf, Switzerland).
For the colony formation assay, 1 × 103 transfected cells per well were seeded in 6-well plates. Next, the cells were incubated at 37°C in the humidified chamber comprising 5% CO2 for 2 weeks. On day 15, the cells were fixed in 4% polyformaldehyde (Sigma‐Aldrich, St. Louis, MO) and rinsed twice with phosphate-buffered saline (PBS). The colonies were then counted (≥50 cells) under a light microscope (Olympus Corporation, Tokyo, Japan).
Cell Apoptosis Quantitation by Flow Cytometry
After 48 h of transfection, the cells were detached using 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.), centrifuged, and washed twice with ice-cold PBS. The proportion of apoptotic cells was determined using the Annexin V/Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (BioLegend, Inc., San Diego, CA, USA). In particular, transfected cells were resuspended in 100 µL of 1× binding buffer. The buffer was then combined with 5 µL of annexin V/FITC and 5 µL of propidium iodide (PI). After 30 min of incubation at room temperature in the dark, the proportion of apoptotic cells was measured in the samples using flow cytometric assays (BD Biosciences, Franklin Lakes, NJ, USA).
Transwell Assay
A total of 100 µL of FBS-free DMEM comprising 5 × 104 cells was added into the upper compartments of transwell inserts that were precoated with Matrigel (both from BD Biosciences), followed by the addition of 600 µL of DMEM comprising 20% FBS into the lower compartments. After culturing for 24 h at 37°C in 5% CO2, the Matrigel and noninvaded cells remaining on the upper surface of the transwell inserts were gently removed with a cotton swab. The invaded cells were fixed in 4% paraformaldehyde, stained with 0.1% crystal violet, and rinsed thrice with PBS. After the random selection of five visual fields (200 × magnification), images of the invaded cells were captured under a light microscope, and the average value was regarded as the number of invaded cells. The migratory capacity of the cells was assessed using an assay similar to the above-mentioned experimental procedure, but the transwell inserts were not coated with Matrigel.
Tumor Xenograft Model
The experimental procedures involving mice were approved by the Animal Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University and conducted in compliance with the 2009 Animal Protection Law of the People’s Republic of China. Male BALB/c nude mice aged 4–6 month and weighing 18–22 g were obtained from Beijing Weitong Lihua Experimental Animal Technology Co., Ltd. (Beijing, China). PCa cells (DU145) transfected with agomir-939 or agomir-NC were subcutaneously inoculated into the flank of nude mice. The width and length of tumor xenografts were measured at 2-day intervals. Four weeks after the injection, all nude mice were euthanized, and the tumor xenografts were obtained for subsequent analysis. Tumor volume was calculated using the following formula: 0.5 × width2 (cm2) × length (cm).
Online Bioinformatics Tools
The targets of miR-939 were searched using microRNA.org (http://www.microrna.org/microrna/home.do) and TargetScan 7.1 (http://www.targetscan.org/).
Luciferase Reporter Assay
The fragments of the HDGF 3′‐UTR comprising wild-type (wt) or mutant (mut) miR-939-binding sites were designed and amplified by GenePharma. The wt and mut fragments were inserted into the psiCHECK‐2™ luciferase plasmid (Promega Corporation, Madison, WI, USA) to generate psiCHECK–HDGF-3′-UTR-wt and psiCHECK–HDGF-3′-UTR-mut reporter plasmids, respectively. Cells were seeded in 24-well plates at 60% confluence. Cotransfection of either psiCHECK–HDGF-3′-UTR-wt or psiCHECK–HDGF-3′-UTR-mut and either agomir-939 or agomir-NC was performed using Lipofectamine™ 2000, according to the manufacturer’s instructions. After cultivation for 48 h, the luciferase activity was evaluated using the Dual Luciferase Reporter Assay Kit (Promega).
Western Blotting
Total protein was extracted with radioimmunoprecipitation assay (RIPA) buffer (Wlaterson, Barcelona, Spain). Total protein concentration was determined with the Bicinchoninic Acid (BCA) Protein Assay Kit (Pierce; Thermo Fisher Scientific, Inc.). The same amounts of protein were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis in 10% polyacrylamide gels and then transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). After blocking with 5% fat-free milk at room temperature for 2 h, the target proteins in the membranes were probed with specific antibodies at 4°C overnight. Thereafter, the membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (ab6721 or ab205719; 1:5000 dilution, Abcam, Cambridge, UK) at room temperature for 2 h. The protein signals were detected using an enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.). The primary antibodies were used at a 1:1000 dilution and included a rabbit monoclonal antihuman HDGF antibody (ab128921; Abcam), rabbit monoclonal antihuman β-catenin antibody (ab32572; Abcam), mouse monoclonal antihuman phospho-(p-)β-catenin antibody (sc-57534; Santa Cruz Biotechnology, Dallas, TX, USA), rabbit monoclonal antihuman cyclin D1 antibody (ab134175; Abcam), and rabbit antihuman GAPDH antibody (sc-32233; Santa Cruz Biotechnology).
Statistical Analysis
All data were expressed as means ± standard deviation and analyzed using the Student’s t-test or one-way analysis of variance, followed by a multiple comparison test. The chi-squared test (χ2 test) was used to determine the association between miR-939 expression and the clinical parameters of patients with PCa. All patients were followed up 100 months, and the Kaplan–Meier method was used to construct the survival curve. Differences among the survival curves were evaluated by the Log rank test. The correlation between miR-939 and HDGF mRNA expression was tested using Spearman correlation analysis. P values of <0.05 were considered statistically significant.
Results
Low miR-939 Expression in PCa Tissues and Cell Lines Correlates with a Poor Prognosis
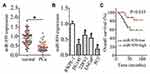
To explore miR-939’s specific characteristics involved in PCa progression, its expression profile was determined in PCa tissues and their corresponding adjacent normal tissues by RT–qPCR. miR-939 expression was found to be lower in the PCa tissues than in their corresponding adjacent normal tissues (Figure 1A, P < 0.05). Consistent with this finding, miR-939 expression was lower in all the tested PCa cell lines—DU145, 22RV1, LNCaP, and PC3—then in the prostatic epithelial cell line RWPE-1 (Figure 1B, P < 0.05).
To determine the clinical value of miR-939 in PCa, all patients with PCa were classified into low miR-939 expression (n = 29) and high miR-939 expression (n = 29) groups. The median value of miR-939 expression among the PCa tissues was defined as the threshold. Then, the correlation between miR-939 expression and the clinical parameters of the patients with PCa was analyzed by the χ2 test. We found that low miR-939 expression was notably correlated with tumor stage (P = 0.033) and lymphatic metastasis (P = 0.028), whereas no significant correlation was observed with age, preoperative prostate-specific antigen level, Gleason score, or distant metastasis (Table 1). In addition, patients with PCa exhibiting low miR-939 expression had a worse overall survival than those with high miR-939 expression (Figure 1C, P = 0.033). These results suggest the strong involvement of miR-939 PCa oncogenicity.
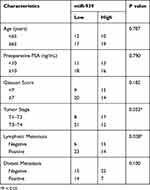 |
Table 1 miR-939 Expression Level (High vs Low) in Correlation with Clinicopathological Characteristics Among Patients with PCa (n=58) |
miR-939 Overexpression Inhibits PCa Cell Proliferation and Colony Formation, Facilitates Apoptosis, and Attenuates Cell Migration and Invasion in vitro
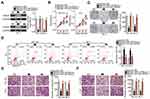
Because of the low miR-939 expression in PCa tissues and cell lines, gain-of-function assays were conducted to evaluate the role of miR-939 in PCa progression via transfection of agomir-939 into DU145 and PC3 cell lines. RT–qPCR findings revealed that compared with the transfection of agomir-NC, the transfection of agomir-939 into DU145 and PC3 cells markedly increased the miR-939 expression (Figure 2A, P < 0.05). The effects of miR-939 overexpression on PCa cell proliferation were examined by CCK-8 and colony formation assays, whereas the effects of miR-939 overexpression on the apoptosis of PCa cell apoptosis were examined using flow cytometric assays. The results revealed that exogenous miR-939 expression suppressed cell proliferation (Figure 2B, P < 0.05) and colony formation (Figure 2C, P < 0.05) and induced apoptosis (Figure 2D, P < 0.05) of DU145 and PC3 cells. Furthermore, transwell assays suggested that compared with agomir-NC-transfected cells, the miR-939-overexpressing DU145 and PC3 cells exhibited impaired migratory (Figure 2E, P < 0.05) and invasive (Figure 2F, P < 0.05) abilities. Taken together, these results suggested that miR-939 overexpression decreased PCa cell growth and metastasis in vitro.
HDGF Is a Direct Target Gene of miR-939 in PCa
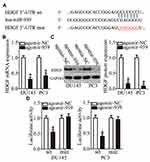
To elucidate the molecular mechanisms underlying the biological functions of miR-939 in PCa, we conducted bioinformatics analysis to predict the potential targets of miR-939 and found a highly conserved binding site for miR-939 in the 3′-UTR of HDGF mRNA (Figure 3A). We next measured HDGF expression in miR-939-overexpressing DU145 and PC3 cells to determine whether miR-939 regulates endogenous HDGF expression. Following exogenous miR-939 overexpression, the mRNA (Figure 3B, P < 0.05) and protein (Figure 3C, P < 0.05) levels of HDGF were significantly decreased in DU145 and PC3 cells. We also performed a luciferase reporter assay to further test our hypothesis that HDGF is a direct target of miR-939 and found that miR-939 upregulation significantly decreased the luciferase activity generated by the plasmid harboring the wt miR-939-binding site (P < 0.05). Conversely, mutation of the binding sequences in the 3′-UTR of HDGF abrogated the suppression of luciferase activity induced by agomir-939 introduction (Figure 3D). These results suggested a direct correlation between miR-939 and the 3′-UTR of HDGF. Taken together, the above results validated HDGF as a direct target of miR-939 in PCa.
HDGF Expression Is Inversely Correlated with miR-939 Expression in PCa Tissues
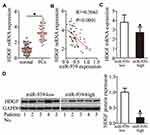
Next, we measured HDGF expression in PCa tissues and their corresponding adjacent normal tissues by RT–qPCR. The expression of HDGF mRNA was significantly higher in PCa tissues than in their corresponding adjacent normal tissues (Figure 4A, P < 0.05). Moreover, there was an inverse correlation between the expression of miR-939 and HDGF mRNA in PCa tissues, as revealed by Spearman correlation analysis (Figure 4B; R2 = 0.3605, P < 0.0001). Furthermore, the mRNA (Figure 4C, P < 0.05) and protein (Figure 4D, P < 0.05) levels of HDGF were lower in the high miR-939 expression group than in the low miR-939 expression group.
Effects of HDGF Knockdown are Similar to Those of miR-939 Upregulation in PCa Cells
To explore the biological roles of HDGF in PCa tumorigenesis, si-HDGF and si-NC were chemically synthesized and introduced into DU145 and PC3 cells. The transfection of si-HDGF clearly downregulated HDGF in DU145 and PC3 cells, as evidenced by Western blotting (Figure 5A, P < 0.05). Next, we examined the influence of HDGF silencing on cell proliferation using CCK-8 and colony formation assays and found that HDGF silencing strongly decreased the proliferative (Figure 5B, P < 0.05) and colony formation (Figure 5C, P < 0.05) abilities of DU145 and PC3 cells. In addition, HDGF knockdown markedly increased the apoptosis rate of DU145 and PC3 cells (Figure 5D, P < 0.05). The results of the transwell assay indicated that inhibition of HDGF significantly attenuated the migration (Figure 5E, P < 0.05) and invasiveness (Figure 5F, P < 0.05) of DU145 and PC3 cells. Taken together, these data suggested that HDGF downregulation can have effects similar to those of miR-939 overexpression in PCa cells, thus confirming HDGF as a downstream mediator of miR-939 in PCa.
HDGF Overexpression Neutralizes the Antitumor Activity of miR-939 in PCa Cells
Rescue experiments were conducted to further confirm that HDGF downregulation is essential for the anticancer activity of miR-939 overexpression in PCa tumorigenesis. To this end, we restored HDGF expression in agomir-939-transfected DU145 and PC3 cells by cotransfecting these cells with the HDGF-overexpressing plasmid pc-HDGF (Figure 6A, P < 0.05). We found that the recovery of HDGF expression countered the effects of miR-939 upregulation on the proliferation (Figure 6B, P < 0.05), colony formation (Figure 6C, P < 0.05), apoptosis (Figure 6D, P < 0.05), migration (Figure 6E, P < 0.05), and invasiveness (Figure 6F, P < 0.05) of DU145 and PC3 cells. These results strongly indicated that HDGF is functionally involved in the miR‐939-mediated suppression of PCa aggressiveness.
miR-939 Overexpression Deactivates the WNT/β-Catenin Pathway in PCa by Downregulating HDGF
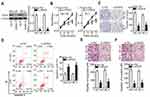
Previous studies have reported the involvement of HDGF in the regulation of the WNT/β-catenin pathway.32–34 Accordingly, we tested whether miR-939 affects WNT/β-catenin signaling in PCa. To this end, we conducted Western blotting to quantify the important molecules associated with the WNT/β-catenin pathway in DU145 and PC3 cells transfected with agomir-939 in the presence of either pc-HDGF or pcDNA3.1. The levels of p-β-catenin and cyclin D1 levels were significantly lower in the agomir-939-transfected cells than in the agomir-NC-transfected cells, whereas total β-catenin expression was unaffected by miR-939 overexpression. Of note, the miR-939 overexpression-mediated decrease in p-β-catenin and cyclin D1 protein expression was reversed in DU145 and PC3 cells after cotransfection with the HDGF-overexpressing plasmid pc-HDGF (Figure 7). These results clearly showed that miR-939 inhibited the activation of the WNT/β-catenin pathway in PCa cells by decreasing HDGF expression.
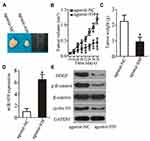
miR-939 Overexpression Impairs Tumor Growth of PCa Cells in vivo
The effect of miR-939 overexpression on the tumor growth of PCa cells in vivo was determined in the tumor xenograft model. DU145 cells were transfected with either agomir-939 or agomir-NC and then inoculated into nude mice to generate a PCa xenograft model. The nude mice injected with miR-939-overexpressing DU145 cells developed remarkably smaller tumor xenografts that exhibited delayed tumor growth (Figure 8A and B, P < 0.05). The tumor xenograft weight was significantly lower in the agomir-939-transfected cells than that in the agomir-NC-transfected cells (Figure 8C, P < 0.05). The successful overexpression of miR-939 was confirmed in the tumor xenografts derived from agomir-939-transfected DU145 cells (Figure 8D, P < 0.05). Western blotting was also performed to determine the expression of HDGF and the components of the WNT/β-catenin pathway in the tumor xenografts. The results revealed that the protein levels of HDGF, p-β-catenin, and cyclin D1 were significantly lower in the agomir-939-transfected cells than in the agomir-NC-transfected cells (Figure 8E). Taken together, our findings suggested that miR-939 overexpression inhibits the tumor growth of PCa cells in vivo by downregulating HDGF and deactivating the WNT/β-catenin pathway.
Discussion
Changes in the expression of miRNAs in PCa cells have been frequently reported.35–37 These abnormally expressed miRNAs may function as key mediators of prostate carcinogenesis and PCa progression by influencing various biological processes.38–40 Therefore, a comprehensive understanding of PCa pathogenesis may improve the diagnosis, prognosis, and treatment of this disease. The present study aimed to determine miR-939 expression in PCa cells and explore its clinical value among patients with PCa. In particular, this study explored the functional roles of miR-939 in PCa progression and the underlying molecular mechanisms.
The expression profile of miR-939 has been well studied in several types of human cancer. For instance, it has been reported that miR-939 is downregulated in tongue squamous cell carcinoma and this downregulation is negatively correlated with tumor stage.41 Low miR-939 expression has also been reported in gastric30 and colorectal29 cancers. This low miR-939 expression is known to be a biomarker for poor prognosis and recurrence in patients with gastric cancer.30 Conversely, miR-939 overexpression has been reported in hepatocellular carcinoma42 and ovarian cancer.31 These conflicting observations increased our interest in determining the expression profile of miR-939 in PCa. The results of the present study revealed that miR-939 was expressed at markedly low levels in PCa tissues and cell lines. Furthermore, clinical analysis showed that this low miR-939 expression was clearly associated with tumor stage and lymphatic metastasis. In addition, patients with PCa exhibiting low miR-939 expression showed poorer prognosis than those exhibiting high miR-939 expression. These results suggested miR-939 as a valuable diagnostic and prognostic indicator of PCa.
The role of miR-939 in tumorigenesis and tumor development has been explored in detail in the literature. For example, restoration of miR-939 expression attenuates the migratory and invasive abilities of colorectal cancer cells by directly targeting LIM domain kinase 2.29 In addition, restored miR-939 expression restricts gastric cancer cell proliferation, migration, and invasion in vitro; increases 5-fluorouracil-induced chemosensitivity; and inhibits lung metastasis in vivo.30 These effects were found to be mediated by a decrease in SLC34A2 expression and deactivation of the RAF/MEK/ERK pathway.30 However, a contradictory function of miR-939 has been observed in ovarian cancer. Ectopic miR-939 expression increases ovarian cancer cell proliferation and anchorage-independent growth by inhibiting the APC2/WNT/β-catenin pathway.31 However, the detailed roles of miR-939 in PCa progression remain elusive. Herein, we found that miR-939 exerted a tumor-suppressive effect on PCa cells by suppressing cell growth and metastasis in vitro and inhibiting tumor growth in vivo.
Detailed investigation of the molecular mechanisms regulating the tumor-suppressive effects of miR-939 on PCa is important to discover novel therapeutic targets and improve the prognosis of patients with PCa. In the present study, HDGF was confirmed as a direct and functional target of miR-939 in PCa. HDGF, located on chromosome 1 region q21–q23, is upregulated in several types of human cancer, including PCa,43,44 endometrial cancer,45 hepatocellular carcinoma,46 ovarian cancer,47 gastric cancer,48 and glioma.49 HDGF serves as an oncogene in the malignant progression of PCa and is implicated in the regulation of cell proliferation, survival, migration, and invasion.43,44 The results of the present study confirmed the oncogenic role of HDGF in PCa progression. Moreover, miR-939 was found to directly target HDGF mRNA to suppress PCa progression via deactivation of the WNT/β-catenin pathway. Accordingly, miR-939 upregulation, which results in HDGF inhibition and WNT/β-catenin pathway deactivation, may be a promising therapeutic strategy for patients with PCa.
Conclusion
The present study characterized the antitumorigenic roles of miR-939 in attenuating the oncogenicity of PCa in vitro and in vivo. In terms of the mechanism, the tumor-suppressive effect of miR-939 on PCa cells is suggested to be mediated by inhibition of the HDGF/WNT/β-catenin pathway. Our study findings regarding miR-939, its direct target gene HDGF, and the WNT/β-catenin pathway involved in PCa may help in identifying novel targets for anticancer therapy.
Abbreviations
3′-UTR, 3′-untranslated region; cDNA, complementary DNA; CCK-8, Cell Counting Kit-8; DMEM, Dulbecco’s Modified Eagle’s Medium; FBS, fetal bovine serum; FITC, fluorescein Isothiocyanate; miRNA, miR, microRNA; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; mut, mutant; NC, negative control; PBS, phosphate-buffered saline; PCa, prostate cancer; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; siRNA, small interfering RNA; wt, wild-type.
Ethics and Consent Statement
All experimental steps involving the use of clinical tissues were approved by the Ethics Committee of Third Affiliated Hospital of Sun Yat-Sen University and were performed in accordance with the Helsinki Declaration. In addition, written informed consent was obtained from all the participating patients. The experimental procedures involving mice were approved by the Animal Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University and were conducted in compliance with the Animal Protection Law of the People’s Republic of China-2009.
Data Sharing Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Author Contributions
Wentao Huang and Jie Situ designed this study. Jie Situ conducted CCK-8 assay, flow cytometry analysis and luciferase reporter assay. Colony formation assay, Transwell assay and RT-qPCR were performed by Hao Zhang and Zi Jin. Ke Li and Yunhua Mao carried out tumor xenograft model and Western blotting. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Science and Technology Planning & Social Development Project of Guangdong Province of China (grant No. 2017A020215027).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
3. Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi:10.3322/CA.2007.0010
4. Heidenreich A, Bastian PJ, Bellmunt J, et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol. 2014;65(1):124–137. doi:10.1016/j.eururo.2013.09.046
5. Egidi MG, Cochetti G, Serva MR, et al. Circulating microRNAs and kallikreins before and after radical prostatectomy: are they really prostate cancer markers? Biomed Res Int. 2013;2013:241780. doi:10.1155/2013/241780
6. Mottet N, Bellmunt J, Bolla M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71(4):618–629. doi:10.1016/j.eururo.2016.08.003
7. Barry MJ, Simmons LH. Prevention of prostate cancer morbidity and mortality: primary prevention and early detection. Med Clin North Am. 2017;101(4):787–806. doi:10.1016/j.mcna.2017.03.009
8. Miyake H, Fujisawa M. Prognostic prediction following radical prostatectomy for prostate cancer using conventional as well as molecular biological approaches. Int J Urol. 2013;20(3):301–311. doi:10.1111/j.1442-2042.2012.03175.x
9. Facina CH, Campos SGP, Goncalves BF, Goes RM, Vilamaior PSL, Taboga SR. Long-term oral exposure to safe dose of bisphenol A in association with high-fat diet stimulate the prostatic lesions in a rodent model for prostate cancer. Prostate. 2018;78(2):152–163. doi:10.1002/pros.23458
10. Poorthuis MHF, Vernooij RWM, van Moorselaar RJA, de Reijke TM. Second-line therapy in patients with metastatic castration-resistant prostate cancer with progression after or under docetaxel: a systematic review of nine randomized controlled trials. Semin Oncol. 2017;44(5):358–371. doi:10.1053/j.seminoncol.2017.10.005
11. Antognelli C, Mezzasoma L, Fettucciari K, Mearini E, Talesa VN. Role of glyoxalase I in the proliferation and apoptosis control of human LNCaP and PC3 prostate cancer cells. Prostate. 2013;73(2):121–132. doi:10.1002/pros.22547
12. Antognelli C, Mezzasoma L, Mearini E, Talesa VN. Glyoxalase 1-419C>A variant is associated with oxidative stress: implications in prostate cancer progression. PLoS One. 2013;8(9):e74014. doi:10.1371/journal.pone.0074014
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
14. Xie B, Ding Q, Han H, Wu D. miRCancer: a microRNA-cancer association database constructed by text mining on literature. Bioinformatics. 2013;29(5):638–644. doi:10.1093/bioinformatics/btt014
15. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
16. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450. doi:10.1016/j.devcel.2006.09.009
17. Pastina P, Nardone V, Croci S, et al. Anti-cancer activity of dose-fractioned mPE ± bevacizumab regimen is paralleled by immune-modulation in advanced squamous NSLC patients. J Thorac Dis. 2017;9(9):3123–3131. doi:10.21037/jtd.2017.08.68
18. Mearini E, Poli G, Cochetti G, Boni A, Egidi MG, Brancorsini S. Expression of urinary miRNAs targeting NLRs inflammasomes in bladder cancer. Onco Targets Ther. 2017;10:2665–2673. doi:10.2147/OTT.S132680
19. Omar HA, El-Serafi AT, Hersi F, et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019;286(18):3540–3557. doi:10.1111/febs.15000
20. Wen X, Deng FM, Wang J. MicroRNAs as predictive biomarkers and therapeutic targets in prostate cancer. Am J Clin Exp Urol. 2014;2(3):219–230.
21. Guelfi G, Cochetti G, Stefanetti V, et al. Next generation sequencing of urine exfoliated cells: an approach of prostate cancer microRNAs research. Sci Rep. 2018;8(1):7111. doi:10.1038/s41598-018-24236-y
22. Poli G, Egidi MG, Cochetti G, Brancorsini S, Mearini E. Relationship between cellular and exosomal miRNAs targeting NOD-like receptors in bladder cancer: preliminary results. Minerva Urol Nefrol. 2019.
23. Zhang X, Jin K, Luo JD, Liu B, Xie LP. MicroRNA-107 inhibits proliferation of prostate cancer cells by targeting cyclin E1. Neoplasma. 2019;66:704–716. doi:10.4149/neo_2018_181105N825
24. Guan C, Zhang L, Wang S, et al. Upregulation of MicroRNA-21 promotes tumorigenesis of prostate cancer cells by targeting KLF5. Cancer Biol Ther. 2019;20:1149–1161.
25. Hong Z, Fu W, Wang Q, Zeng Y, Qi L. MicroRNA-384 is lowly expressed in human prostate cancer cells and has anti-tumor functions by acting on HOXB7. Biomed Pharmacother. 2019;114:108822. doi:10.1016/j.biopha.2019.108822
26. Cochetti G, Poli G, Guelfi G, Boni A, Egidi MG, Mearini E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016;9:7545–7553. doi:10.2147/OTT.S119027
27. Kanwal R, Plaga AR, Liu X, Shukla GC, Gupta S. MicroRNAs in prostate cancer: functional role as biomarkers. Cancer Lett. 2017;407:9–20. doi:10.1016/j.canlet.2017.08.011
28. Egidi MG, Cochetti G, Guelfi G, et al. Stability assessment of candidate reference genes in urine sediment of prostate cancer patients for miRNA applications. Dis Markers. 2015;2015:973597. doi:10.1155/2015/973597
29. Zhang Y, Liu X, Li Q, Zhang Y. lncRNA LINC00460 promoted colorectal cancer cells metastasis via miR-939-5p sponging. Cancer Manag Res. 2019;11:1779–1789. doi:10.2147/CMAR.S192452
30. Zhang JX, Xu Y, Gao Y, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16(1):18. doi:10.1186/s12943-017-0586-y
31. Ying X, Li-ya Q, Feng Z, Yin W, Ji-hong L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed Pharmacother. 2015;71:64–69. doi:10.1016/j.biopha.2015.02.020
32. Zheng Y, Lu S, Xu Y, Zheng J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/beta-catenin signaling pathway. Int J Biol Macromol. 2019;128:521–530. doi:10.1016/j.ijbiomac.2019.01.121
33. Liu C, Wang L, Jiang Q, et al. Hepatoma-derived growth factor and DDX5 promote carcinogenesis and progression of endometrial cancer by activating beta-catenin. Front Oncol. 2019;9:211. doi:10.3389/fonc.2019.00211
34. Lian J, Tang J, Shi H, et al. Positive feedback loop of hepatoma-derived growth factor and beta-catenin promotes carcinogenesis of colorectal cancer. Oncotarget. 2015;6(30):29357–29374. doi:10.18632/oncotarget.4982
35. Zhang FB, Du Y, Tian Y, Ji ZG, Yang PQ. MiR-1299 functions as a tumor suppressor to inhibit the proliferation and metastasis of prostate cancer by targeting NEK2. Eur Rev Med Pharmacol Sci. 2019;23(2):530–538. doi:10.26355/eurrev_201901_16865
36. Zhang X, Zhou J, Xue D, Li Z, Liu Y, Dong L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. doi:10.1016/j.ijbiomac.2019.01.127
37. Zhao H, Lai X, Zhang W, et al. MiR-30a-5p frequently downregulated in prostate cancer inhibits cell proliferation via targeting PCLAF. Artif Cells Nanomed Biotechnol. 2019;47(1):278–289. doi:10.1080/21691401.2018.1553783
38. Aghdam SG, Ebrazeh M, Hemmatzadeh M, et al. The role of microRNAs in prostate cancer migration, invasion, and metastasis. J Cell Physiol. 2019;234(7):9927–9942. doi:10.1002/jcp.27948
39. Sharma N, Baruah MM. The microRNA signatures: aberrantly expressed miRNAs in prostate cancer. Clin Transl Oncol. 2019;21(2):126–144. doi:10.1007/s12094-018-1910-8
40. Moustafa AA, Kim H, Albeltagy RS, El-Habit OH, Abdel-Mageed AB. MicroRNAs in prostate cancer: from function to biomarker discovery. Exp Biol Med. 2018;243(10):817–825. doi:10.1177/1535370218775657
41. Chen Y, Guo Y, Yan W. lncRNA RP5-916L7.2 correlates with advanced tumor stage, and promotes cells proliferation while inhibits cells apoptosis through targeting miR-328 and miR-939 in tongue squamous cell carcinoma. Clin Biochem. 2019;67:24–32. doi:10.1016/j.clinbiochem.2019.02.013
42. Fornari F, Ferracin M, Trere D, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10(10):e0141448. doi:10.1371/journal.pone.0141448
43. Shetty A, Dasari S, Banerjee S, et al. Hepatoma-derived growth factor: a survival-related protein in prostate oncogenesis and a potential target for vitamin K2. Urol Oncol. 2016;34(11):483e481–483 e488. doi:10.1016/j.urolonc.2016.05.027
44. Guo Z, He Y, Wang S, et al. Various effects of hepatoma-derived growth factor on cell growth, migration and invasion of breast cancer and prostate cancer cells. Oncol Rep. 2011;26(2):511–517. doi:10.3892/or.2011.1295
45. Wang L, Jiang Q, Hua S, et al. High nuclear expression of HDGF correlates with disease progression and poor prognosis in human endometrial carcinoma. Dis Markers. 2014;2014:298795. doi:10.1155/2014/298795
46. Chen S-C, Hu T-H, Huang C-C, et al. Hepatoma-derived growth factor/nucleolin axis as a novel oncogenic pathway in liver carcinogenesis. Oncotarget. 2015;6(18):16253–16270. doi:10.18632/oncotarget.3608
47. Liu XJ, Liu WL, Yang FM, Yang XQ, Lu XF. Hepatoma-derived growth factor predicts unfavorable prognosis of epithelial ovarian cancer. Onco Targets Ther. 2015;8:2101–2109. doi:10.2147/OTT.S85660
48. Yamamoto S, Tomita Y, Hoshida Y, et al. Expression of hepatoma-derived growth factor is correlated with lymph node metastasis and prognosis of gastric carcinoma. Clin Cancer Res. 2006;12(1):117–122. doi:10.1158/1078-0432.CCR-05-1347
49. Yang Y, Liang S, Li Y, et al. Hepatoma-derived growth factor functions as an unfavorable prognostic marker of human gliomas. Oncol Lett. 2017;14(6):7179–7184. doi:10.3892/ol.2017.7180
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.