Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-769-5p Inhibits Pancreatic Ductal Adenocarcinoma Progression by Directly Targeting and Downregulating ETS Proto-Oncogene 1
Authors Cheng K, Feng L, Yu S, Yu C, Chi N
Received 10 June 2019
Accepted for publication 7 December 2019
Published 3 January 2020 Volume 2019:12 Pages 11737—11750
DOI https://doi.org/10.2147/OTT.S218876
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Tohru Yamada
This paper has been retracted.
Kai Cheng, 1 Lan Feng, 2 Shuang Yu, 3 Changhong Yu, 1 Nannan Chi 1
1Department of Gastroenterology, First Affiliated Hospital of Jiamusi University, Jiamusi, Heilongjiang 154002, People’s Republic of China; 2Department of Infectious Diseases, First Affiliated Hospital of Jiamusi University, Jiamusi, Heilongjiang 154002, People’s Republic of China; 3Department of Cardiovascular Medicine, First Affiliated Hospital of Jiamusi University, Jiamusi, Heilongjiang 154002, People’s Republic of China
Correspondence: Nannan Chi
Department of Gastroenterology, First Affiliated Hospital of Jiamusi University, No. 348 Dexiang Road, Jiamusi, Heilongjiang 154002, People’s Republic of China
Email [email protected]
Purpose: MicroRNA-769-5p (miR-769) is aberrantly expressed and plays crucial roles in non–small cell lung cancer and melanoma. However, the expression pattern, biological role, and mechanisms of action of miR-769 in pancreatic ductal adenocarcinoma (PDAC) are yet to be fully elucidated. Therefore, we attempted to determine the potential regulatory function of miR-769 in PDAC progression and to explore the underlying mechanisms in detail.
Methods: In this study, reverse-transcription quantitative polymerase chain reaction was carried out to determine the expression profile of miR-769 in PDAC. A series of experiments, including a Cell Counting Kit-8 assay, flow-cytometric analysis, Transwell migration and invasion assays, and a xenograft animal model, were applied to test whether miR-769 affects the malignancy of PDAC.
Results: We found that miR-769 was significantly underexpressed in PDAC tissues and cell lines. The low miR-769 expression significantly correlated with the TNM stage and lymph node metastasis. Patients with PDAC harboring low miR-769 expression showed shorter overall survival than did the patients with high miR-769 expression. Forced upregulation of miR-769 suppressed PDAC cell proliferation, migration, and invasion in vitro; promoted apoptosis in vitro; and hindered tumor growth in vivo. Experiments on the mechanism identified ETS proto-oncogene 1 (ETS1) as a direct target gene of miR-769 in PDAC cells. Furthermore, ETS1 turned out to be upregulated in PDAC tissue samples, and the upregulation of ETS1 negatively correlated with miR-769 expression. Moreover, ETS1 knockdown simulated the tumor-suppressive effects of miR-769 overexpression on PDAC cells. Besides, ETS1 reintroduction attenuated the antitumor actions of miR-769 upregulation in PDAC cells.
Conclusion: Our findings indicate that miR-769 performs tumor-suppressive functions in PDAC by directly targeting ETS1, and this miRNA may represent a potential therapeutic target for the development of anticancer therapies.
Keywords: pancreatic ductal adenocarcinoma, microRNA-769-5p, proliferation, invasion, ETS proto-oncogene 1
Introduction
Pancreatic cancer ranks the fifth most common cancer and the second leading cause of cancer-associated deaths around the world.1 Pancreatic ductal adenocarcinoma (PDAC), the most common subtype of pancreatic cancer, accounts for approximately 90% of all pancreatic cancer cases.2 Even though great research efforts have been devoted to the diagnosis and treatments in the last two decades, the clinical outcomes of patients with PDAC remain quite poor, with a 5-year survival rate of less than 7%.3 The poor prognosis of PDAC is mainly associated with the fact that PDACs are prone to local infiltration, metastasis, and recurrence even after surgical resection.4,5 In recent years, remarkable development has been made in understanding the mechanisms underlying the malignant behaviors of PDAC, but the detailed pathogenesis is still largely unknown and needs further research.6 Therefore, exploration of the mechanisms implicated in the regulation of PDAC initiation and progression is urgently needed for the identification of effective methods for the treatment of patients with this fatal disease.
MicroRNAs (miRNAs) are a group of endogenous noncoding short RNA molecules with a length of 18–22 nucleotides.7 They negatively regulate gene expression by directly binding to the 3′-untranslated region (3′-UTR) of their target mRNAs in a sequence-specific manner.8 This binding event causes mRNA degradation and/or translational suppression, thereby reducing protein expression.9 It is well documented that miRNAs are aberrantly expressed and participate in the pathogenesis of various human disorders, including cancer.10–12 The dysregulation of miRNAs has been detected in nearly all human cancer types, and may play either tumor-suppressive or oncogenic roles, which mainly depend on the characteristics of their target genes.13–15 Many miRNAs have previously been reported to be downregulated or upregulated in PDAC, and their deregulation is involved in the control of cancer-related biological behaviors, including cell proliferation, cell cycle distribution, apoptosis, metastasis, and resistance to radiotherapy and chemotherapy.16–18 Thus, elucidating the detailed roles and associated mechanisms of action of deregulated miRNAs in PDAC will be particularly useful for the development of diagnostic biomarkers and therapeutic targets.
ETS1 belongs to the ETS family of transcription factors that share a unique DNA binding domain.19 It is a 54 kDa nuclear protein and implicated in the tumorigenesis and tumor development.20 ETS1 results a great deal of tumor behavior, including rell proliferation, cell cycle, apoptosis, angiogenesis, migration, invasion and epithelial-mesenchymal transition.21–23 Previous studies have revealed that ETS1 could be directly targeted and negatively regulated by miRNAs in human cancers. For instance, miR-127 suppresses the malignant phenptype of prostate cancer through directly targeting ETS1 and inhibiting the PI3K/AKT/mTOR Pathway.24 MiR-139-5p directly targets ETS1 to restrict the aerobic Glycolysis, proliferation, metastasis of hepatocellular carcinoma.25 However, the cross-talk between ETS1 and miRNAs in PDAC is yet to be elucidated. MiR-769-5p (miR-769) has attracted our attention owing to its significant participation in the regulation of cancer biology of non–small cell lung cancer26 and melanoma.27 Nonetheless, the expression pattern, biological function, and the mechanisms of action of miR-769 in PDAC are yet to be fully elucidated. Hence, in our study, we aimed to detect miR-769 expression in PDAC tissues and cell lines and to determine the potential regulatory involvement of miR-769 in PDAC progression. In addition, the mechanisms of action of miR-769 in PDAC cells were explored. The discovery of the involvement of miR-769 in PDAC may provide novel insight into the molecular mechanisms and treatment of PDAC.
Materials and Methods
Tissue Collection
PDAC tissues and matched adjacent normal pancreatic tissues were collected from 53 patients with PDAC who underwent surgical resection at the First Affiliated Hospital of Jiamusi University. None of the patients had been treated with chemotherapy or radiotherapy before surgical resection. All tissue samples were rapidly frozen in liquid nitrogen and then stored at –80°C. This study was approved by the Ethics Committee of the First Affiliated Hospital of Jiamusi University and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all the participants regarding the use of clinical samples for study purposes.
Cell Culture and Transfection
Normal human pancreatic cell line HPDE6c7 was bought from the American Type Culture Collection (Manassas, VA, USA). Four PDAC cell lines, Sw1990, Bxpc-3, Panc-1, and Aspc-1, were ordered from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All the cell lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% of fetal bovine serum (FBS), 100 U/mL penicillin, and 100 µg/mL streptomycin (all from Gibco, Thermo Scientific, Shanghai, China). The cells were routinely cultured at 37°C in an atmosphere containing 5% of CO2.
To restore miR-769 expression, the cells were seeded in 6-well plates at a density of 8 × 105 cells/well and were transfected with miR-769 mimics (Shanghai GenePharma Co., Ltd., Shanghai, China). The miRNA mimics negative control (miR-NC) was also purchased from Shanghai GenePharma Co., Ltd., and served as an internal control for the miR-769 mimics. Small interfering RNA (siRNA) used to silence ETS1 (si-ETS1) and si-NC were provided by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). An ETS1 overexpression plasmid lacking its 3′-UTR (pcDNA3.1-ETS1) and the empty pcDNA3.1 vector were chemically synthesized by the Chinese Academy of Sciences (Changchun, China). Plasmid pcDNA3.1-ETS1 was introduced into cells to upregulate endogenous ETS1 expression, with the empty pcDNA3.1 vector as an internal control. All transfection procedures were performed using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) in accordance with the manufacturer’s instructions.
Reverse-Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was isolated from tissues or cells with the TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer’s protocol. For analysis of miR-769 expression, total RNA was subjected to cDNA synthesis using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). After that, qPCR was carried out with the TaqMan MicroRNA PCR Kit (Applied Biosystems). To measure ETS1 mRNA expression, cDNA was chemically synthesized from total RNA through reverse transcription with M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA). After that, the expression level of ETS1 mRNA was assessed using the SYBR-Green Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). Small nuclear RNA U6 and GAPDH served as controls for the normalization of miR-769 and ETS1 mRNA, respectively. Relative gene expression was calculated by the 2−ΔΔCq method.28
Cell Counting Kit-8 (CCK-8) Assay
Following 24 h of transfection, 3000 cells per well were seeded in 96-well culture plates and were maintained at 37°C in an atmosphere supplied with 5% of CO2 for different periods: 0, 24, 48, and 72 h after inoculation. Cellular proliferation was evaluated with the CCK-8 assay (Dojindo, Kumamoto, Japan). Briefly, 10 µL of the CCK-8 solution was added into each well followed by incubation at 37°C for additional 2 h. The optical density (OD) of each well was detected at a 450 nm wavelength on a spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Flow-Cytometric Analysis for Evaluation of Apoptosis
The detection of apoptosis was performed using the Annexin V-Fluorescein Isothiocyanate (FITC) Apoptosis Detection Kit (Biolegend, San Diego, CA, USA). The transfected cells were harvested after 48 h of incubation, rinsed thrice with ice-cold phosphate buffer and resuspended in 100 µL of binding buffer. Next, the cell suspension was stained with 5 µL of Annexin V-FITC and 5 µL of the propidium iodide solution that came with the kit. The percentage of apoptotic cells was determined by means of a flow cytometer (FACScan™; BD Biosciences, Franklin Lakes, NJ, USA) after 20 min incubation at room temperature in darkness.
Transwell Migration and Invasion Assays
For the invasion assay, 105 transfected cells in 200 µL of FBS-free DMEM were plated into the top compartment of Transwell chambers precoated with Matrigel (both from BD Biosciences). The lower compartments were covered with 500 µL of DMEM containing 20% of FBS to act as a chemoattractant. After cultivation for 24 h, noninvading cells that remained on the upper side of the Transwell chamber were gently removed with a cotton swab. The invading cells were fixed with 100% methanol, stained with 0.1% crystal violet, and quantified. The invading cells in five randomly selected visual fields were counted under an inverted light microscope (Olympus Corporation, Tokyo, Japan). The Transwell migration assay was carried out in the same way as the invasion assay was, except that the chambers were not precoated with Matrigel.
Xenograft Animal Model
The procedures of all animal experiments were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Jiamusi University and were conducted following the Animal Protection Law of the People’s Republic of China-2009. Female BALB/c nude mice were bought from the Animal Center of the Second Military Medical University (Shanghai, China). The miR-769 mimics–transfected or miR-NC–transfected Sw1990 cells were subcutaneously injection into the flank of nude mouse. At 2 weeks after the inoculation, the width and length of tumor xenografts were measured using vernier calipers. Tumor volumes were calculated via the following formula: Volume (mm3) = 0.5 × width2 (mm2) × length (mm). The measurement of tumor volume was performed every 2 days until 4 weeks after injection. All nude mice were euthanized to excise the tumor xenografts, and the tumor weight was measured.
Immunohistochemistry
The fixation of tumor xenografts in 4% neutral formalin was conducted at room temperature for 24 h, after which was then embedded in paraffin. After the paraffin is cut into 4 μm sections, they were deparaffinized in xylene, rehydrated in a graded alcohol series and probed with boiling citrate buffer. In order to decrease the non-specific binding, the sections were probed with 5% bovine serum albumin (R&D Systems, Minneapolis, MN, USA) at 37 °C for 45 min. After 16 h incubation with Ki-67 antibody (sc-23900; Santa Cruz Biotechnology, Dallas, TX, USA) at 4 °C, the sections were further incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. Thereafter, the sections were counterstained with 1% hematoxylin at room temperature for 5 min, followed by dehydration in a graded series of ethanol. Finally,the iamges were capatured under an light microscope.
Identification of miR-769 Target Genes
Bioinformatics tools, including TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/), and miRanda (http://www.microrna.org), were utilized to search for the potential target genes of miR-769.
Luciferase Reporter Assay
The wild-type (wt) 3′-UTR fragment of ETS1 containing the predicted miR-769–binding sequences and the mutant 3′-UTR fragment were chemically synthesized by Shanghai GenePharma Co., Ltd., and cloned into the pmirGLO dual-luciferase miRNA target expression vector (Promega) to generate the pmirGLO-ETS1-3ʹ-UTR wt and pmirGLO-ETS1-3ʹ-UTR mut, respectively. For the reporter assay, cells were seeded in 24-well plates at a density of 5 × 105 cells per well and were cotransfected with either the miR-769 mimics or miR-NC and either pmirGLO-ETS1-3ʹ-UTR wt or pmirGLO-ETS1-3ʹ-UTR mut by means of Lipofectamine 2000. Luciferase activity was measured 48 h after transfection using a Dual-Luciferase Reporter Assay System (Promega) as per the manufacturer’s protocol. The activity of firefly luciferase was normalized to that of Renilla luciferase.
Western Blot Analysis
The extraction of total protein from cultured cells was performed with RIPA lysis buffer (Solarbio, Beijing, China). The concentration of total protein was measured with the BCA Protein Assay Kit (Beyotime, Shanghai, China). The equivalent amounts of total protein were separated by SDS polyacrylamide gel electrophoresis in a 10% gel and transferred to polyvinylidene difluoride membranes, which were then blocked at room temperature for 2 h in 5% fat-free milk diluted in Tris-buffered saline with 0.1% of Tween 20 (TBST). After overnight incubation at 4°C with primary antibodies in TBST, the membranes were washed with TBST thrice and then incubated with a horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (sc-516102; 1:5000 dilution; Santa Cruz Biotechnology, Dallas, TX, USA) at room temperature for 2 h. The detection of protein signals was carried out by means of an Enhanced Chemiluminescence Detection System (Pierce, Rockford, IL, USA). The primary antibodies used in this study included a mouse anti-human ETS1 monoclonal antibody (sc-55581; 1:1000 dilution; Santa Cruz Biotechnology), and a mouse anti-human GAPDH antibody (sc-47724; 1:1000 dilution; Santa Cruz Biotechnology). GAPDH served as a loading control.
Statistical Analysis
All data are presented as mean ± standard deviation. Statistical analysis was performed in the SPSS software, version 16.0 (SPSS, Chicago, IL, USA). The differences between groups were evaluated by Student’s t-test or one-way analysis of variance followed by the Student–Newman–Keuls post hoc test. Survival analysis was conducted by the Kaplan–Meier method and logrank test. The chi-square test was carried out for analysis of the possible correlation between miR-769 expression and clinical parameters among the patients with PDAC. The association between miR-769 and ETS1 mRNA levels was assessed by Spearman correlation analysis. Data with P<0.05 were considered statistically significant.
Results
miR-769 Expression Levels are Reduced in PDAC Tissues and Cell Lines
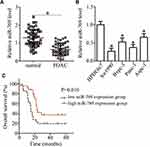
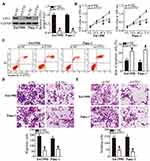
To uncover the expression pattern of miR-769 in PDAC, we first analyzed miR-769 expression in 53 pairs of PDAC tissue samples and matched adjacent normal pancreatic tissue samples by RT-qPCR. The expression of miR-769 was lower in PDAC tissue samples than in adjacent normal pancreatic tissues (Figure 1A, P<0.05). Additionally, miR-769 expression was measured in four PDAC cell lines (Sw1990, Bxpc-3, Panc-1, and Aspc-1) and a normal human pancreatic cell line, HPDE6c7. The results obtained in the RT-qPCR analysis indicated that miR-769 was downregulated in all four tested PDAC cell lines compared with HPDE6c7 cells (Figure 1B, P<0.05).
We then determined the clinical value of the decreased miR-769 expression in patients with PDAC. According to the median value of miR-769 expression among PDAC tissue samples, all patients were classified into two subtypes: the low miR-769 expression group (n = 27) and high miR-769 expression group (n = 26). Analysis of correlation between miR-769 expression and clinical parameters revealed that the reduced miR-769 expression notably correlated with the TNM stage (P=0.027) and lymph node metastasis (P=0.011; Table 1). Furthermore, Kaplan–Meier survival analysis indicated that overall survival of the patients with PDAC harboring low miR-769 expression was obviously shorter than that of patients with high miR-769 expression (Figure 1C, P=0.010). These results suggested that reduced miR-769 levels may be closely related to the progression of PDAC.
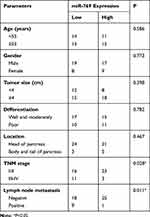 |
Table 1 The Correlation Between miR-769 Expression and Clinical Parameters Among Patients with PDAC |
miR-769 Upregulation Inhibits the Growth and Metastasis of PDAC Cells in vitro
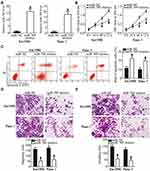
The expression of miR-769 was relatively lower in Sw1990 and Panc-1 cell lines than in Bxpc-3 and Aspc-1 cell lines. Therefore, the two cell lines were selected for further experiments. To investigate the potential biological role of miR-769 in PDAC, Sw1990 and Panc-1 cell lines were transfected with the miR-769 mimics or miR-NC. As a result, miR-769 was effectively overexpressed in Sw1990 and Panc-1 cells after transfection with the miR-769 mimics (Figure 2A, P<0.05). The CCK-8 assay and flow cytometry were performed to determine the role of miR-769 upregulation in PDAC cell proliferation and apoptosis in vitro. It was observed that Sw1990 and Panc-1 cells transfected with the miR-769 mimics showed reduced proliferation (Figure 2B, P<0.05) and increased apoptosis (Figure 2C, P<0.05) as compared with the cells transfected with miR-NC. Next, to evaluate the effect of miR-769 on the metastasis of PDAC cells, Transwell migration and invasion assays were conducted on Sw1990 and Panc-1 cells after miR-769 mimics or miR-NC transfection. The results revealed that exogenous miR-769 expression significantly decreased the migratory (Figure 2D, P<0.05) and invasive (Figure 2E, P<0.05) abilities of Sw1990 and Panc-1 cells. Altogether, these results implied that miR-769 may play a tumor-suppressive part in the progression of PDAC by inhibiting cell growth and metastasis in vitro.
miR-769 Directly Binds to the 3′-UTR of ETS1 mRNA and Inhibits ETS1 Expression in PDAC Cells
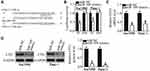
To elucidate the molecular mechanism by which miR-769 attenuated the aggressiveness of PDAC cells, bioinformatics analysis was performed to predict the potential target of miR-769. The 3′-UTR of ETS1 contains a putative binding site for miR-769 (Figure 3A) and was chosen for verification because this gene has previously been implicated in the malignant progression of PDAC.29–33 To test whether miR-769 can directly bind to the 3′-UTR of ETS1 mRNA, the luciferase reporter assay was performed on Sw1990 and Panc-1 cells that were cotransfected with either the miR-769 mimics or miR-NC and either pmirGLO-ETS1-3ʹ-UTR wt or pmirGLO-ETS1-3ʹ-UTR mut. The results revealed that, compared with the miR-NC group, upregulation of miR-769 obviously decreased the luciferase activity of the plasmid carrying the wild-type miR-769–binding site in the 3′-UTR of ETS1 (P<0.05). By contrast, the suppressive effects were abrogated in Sw1990 and Panc-1 cells transfected with a reporter plasmid containing the mutant miR-769–binding site (Figure 3B). Next, we assessed the regulatory influence of miR-769 on endogenous ETS1 expression in PDAC cells. As predicted, forced miR-769 expression decreased ETS1 expression at both mRNA (Figure 3C, P<0.05) and protein levels (Figure 3D, P<0.05) in Sw1990 and Panc-1 cells. Overall, these results meant that ETS1 is a direct target gene of miR-769 in PDAC cells. This finding was consistent with our hypothesis.
miR-769 Inversely Correlates with ETS1 Levels in PDAC Tissues
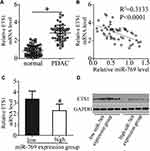
ETS1 was validated as a direct target gene of miR-769 in PDAC cells; hence, we then attempted to further evaluate the relation between miR-769 and ETS1 in PDAC tissues. First, RT-qPCR was performed to determine ETS1 mRNA levels in 53 pairs of PDAC tissue samples and matched adjacent normal pancreatic tissues. The expression of ETS1 mRNA was noticeably higher in PDAC tissue samples than in normal pancreatic tissue samples (Figure 4A, P<0.05). In addition, an inverse association was noted between miR-769 and ETS1 mRNA levels among PDAC tissue samples (Figure 4B; R2 = 0.3133, P<0.0001). Furthermore, the mRNA (Figure 4C, P<0.05) and protein levels (Figure 4D, P<0.05) of ETS1 were obviously lower in the high miR-769 expression group than in the low miR-769 expression group. These results indicated that the upregulation of ETS1 in PDAC tissue samples, at least partly, was caused by miR-769 underexpression.
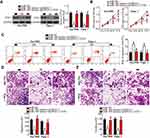
Silencing of ETS1 Expression Simulates the Tumor-Suppressive Effects of miR-769 Overexpression on PDAC Cells
Having identified ETS1 as a direct target of miR-769, next, we illustrated the involvement of ETS1 in the malignant phenotype of PDAC in detail. Loss-of-function assays were performed via introduction of si-ETS1 into Sw1990 and Panc-1 cells. Transfection of si-ETS1 efficiently knocked down ETS1 expression in Sw1990 and Panc-1 cells, as evidenced by Western blotting (Figure 5A, P<0.05). Then, functional experiments indicated that ETS1 knockdown in the Sw1990 and Panc-1 cells reduced their proliferation (Figure 5B, P<0.05) and promoted their apoptosis (Figure 5C, P<0.05) in vitro. Moreover, the migration (Figure 5D, P<0.05) and invasiveness (Figure 5E, P<0.05) of Sw1990 and Panc-1 cells were markedly impaired after the ETS1 knockdown. Therefore, these observations confirmed that ETS1 downregulation mediated the effects of miR-769 in PDAC cells.
Overexpression of ETS1 Attenuates the Anticancer Effects of miR-769 on PDAC Cells
A series of rescue experiments was carried out to verify whether the tumor-suppressive activity of miR-769 in PDAC cells is mediated by the repression of ETS1. Sw1990 and Panc-1 cells were cotransfected with the miR-769 mimics and either ETS1-overexpressing plasmid pcDNA3.1-ETS1 or empty vector pcDNA3.1. Western blot analysis proved that the ETS1 protein level recovered in Sw1990 and Panc-1 cells cotransfected with the miR-769 mimics and pcDNA3.1-ETS1 relative to that in the cells cotransfected with the miR-769 mimics and empty pcDNA3.1 (Figure 6A, P<0.05). In functional analysis, reintroduction of ETS1 expression partially reversed the influence of miR-769 on Sw1990 and Panc-1 cell proliferation (Figure 6B, P<0.05), apoptosis (Figure 6C, P<0.05), migration (Figure 6D, P<0.05), and invasion (Figure 6E, P<0.05). Accordingly, these results clearly indicated that the inhibitory actions of miR-769 on PDAC cell growth and metastasis at least in part were mediated by direct inhibition of ETS1 expression.
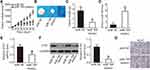
miR-769 Overexpression Hinders PDAC Tumor Growth in vivo
To examine the impact of miR-769 on tumor growth in vivo, Sw1990 cells transfected with the miR-769 mimics or miR-NC were subcutaneously inoculated into the flank of nude mice to establish the xenograft model. The nude mice injected with miR-769–overexpressing Sw1990 cells developed obviously smaller tumor xenografts than did the miR-NC group (Figure 7A and B, P<0.05). In addition, the tumor weight was significantly lower in the miR-769 mimics group than in the miR-NC group (Figure 7C, P<0.05). Furthermore, the successful overexpression of miR-769 (Figure 7D, P<0.05) and downregulation of ETS1 mRNA (Figure 7E, P<0.05) and protein levels (Figure 7F, P<0.05) were validated in the tumor xenografts derived from the miR-769 mimics group. Then, immunohistochemistry results indicated that tumor xenograft overexpressing miR-769 displayed lower Ki-67 expression compared with that in miR-NC group (Figure 7G). All these results suggested that miR-769 overexpression decreased PDAC growth in vivo by reducing ETS1 expression.
Discussion
Many miRNAs have been reported to be deregulated in PDAC, and the deregulation of miRNAs has been implicated in the carcinogenesis and progression of PDAC.34–36 Therefore, better knowledge about miRNAs in PDAC may improve our understanding of the PDAC pathogenesis and facilitate the identification of valuable therapeutic targets in this aggressive disease. Nevertheless, the detailed roles of specific miRNAs in PDAC still remain largely undefined. Here, for the first time, we attempted to measure miR-769 expression in PDAC and to determine the biological functions of miR-769 in the progression of PDAC. Moreover, the mechanisms underlying the actions of miR-769 in PDAC cells were explored.
MiR-769 is downregulated in non–small cell lung cancer, and this downregulation is significantly associated with the clinical stage and lymph node metastasis.26 Patients with non–small cell lung cancer harboring low miR-769 levels show poorer clinical outcomes than do patients with high miR-769 levels.26 In contrast, miR-769 is upregulated in melanoma.27 These conflicting findings indicate that the human miR-769 expression pattern is cancer-specific. However, the expression status of miR-769 in PDAC has been unknown. In our study, we measured its expression in PDAC tissue samples and cell lines by RT-qPCR. Our results indicate that miR-769 is downregulated in PDAC, and its downregulation is associated with TNM stage and lymph node metastasis. Furthermore, the overall survival of patients with PDAC harboring low miR-769 expression was obviously shorter than that of patients with high miR-769 expression. Hence, miR-769 might be an attractive diagnostic and/or prognostic biomarker of PDAC.
MiR-769 has a tumor suppressor activity in non–small cell lung cancer. MiR-769 directly targets TGFBR1 mRNA to inhibit the growth and metastasis of non–small cell lung cancer cells both in vitro and in vivo.26 By contrast, miR-769 is identified as an oncogenic miRNA in melanoma. Upregulation of miR-769 promotes melanoma cell growth and colony formation in vitro by directly targeting GSK3B mRNA.27 However, whether miR-769 is involved in the regulation of PDAC progression has remained largely elusive. This study revealed that restoration of miR-769 expression works as a tumor suppressor in PDAC by inhibiting cell growth and metastasis in vitro and tumor growth in vivo. These findings suggest that miR-769 might be validated as a potential target for anticancer therapies against PDAC.
Identification of the direct target genes of miR-769 in PDAC is important for understanding the participation of miR-769 in the pathogenesis and progression of PDAC. Hence, a series of experiments was conducted to investigate the direct target genes of miR-769 in PDAC cells. ETS1, a member of the ETS family of transcription factors, was validated as a direct target of miR-769 in PDAC cells. ETS1 directly interacts with the specific DNA sequences containing the GGAA/T core motif and participates in cancer formation and progression. It is upregulated in various types of human cancer, such as colorectal cancer,37 breast cancer,38 gastric cancer,39 and laryngeal squamous cell carcinoma.40 Besides, ETS1 is overexpressed in PDAC, and the upregulation of ETS1 is related to tumor differentiation status.29 ETS1 plays crucial roles in the malignant progression of PDAC cells by regulating cell proliferation, metastasis, epithelial–mesenchymal transition, drug resistance, and angiogenesis.30–33 In our current study, we demonstrated that miR-769 directly targets ETS1 mRNA to inhibit the malignancy of PDAC cells in vitro and in vivo. Thus, miR-769–mediated inhibition of ETS1 might be a potential therapeutic strategy against PDAC, to be validated in the future.
There is a limitation of our study. In this study, we did not investigate the effect of miR-769 overexpression on the metastasis of PDAC cells in vivo. In our further investigations, miR-769 mimics and miR-NC-transfected PDAC cells will be injected into the tail vein of nude mice, and utilized for determining the capacity of PDAC cells to extravasate and grow at metastatic sites.
Conclusion
This study revealed that miR-769 exerts tumor-suppressive actions in PDAC, at least partly by directly targeting ETS1 mRNA. These results form a theoretical basis for the application of miR-769 to the treatment of PDAC.
Abbreviations
3′-UTR, 3′-untranslated region; CCK-8, Cell Counting Kit-8; DMEM, Dulbecco’s Modified Eagle’s Medium; FBS, fetal bovine serum; FITC, fluorescein Isothiocyanate; miRNA, miR, microRNA; mut, mutant; NC, negative control; PDAC, pancreatic ductal adenocarcinoma; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; TBST, Tris-buffered saline with 0.1% of Tween 20; wt, wild-type.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of the First Affiliated Hospital of Jiamusi University and was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all the participants regarding the use of clinical samples for study purposes. The procedures of all animal experiments were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Jiamusi University and were conducted following the Animal Protection Law of the People’s Republic of China-2009.
Data Sharing Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Acknowledgments
The study was supported by Natural Science Foundation of Heilongjiang Province (H2016088). Kai Cheng and Lan Feng are co-first authors for this study.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Subramani R, Gangwani L, Nandy SB, Arumugam A, Chattopadhyay M, Lakshmanaswamy R. Emerging roles of microRNAs in pancreatic cancer diagnosis, therapy and prognosis (Review). Int J Oncol. 2015;47(4):1203–1210. doi:10.3892/ijo.2015.3129
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254
4. Campa D, Rizzato C, Capurso G, et al. Genetic susceptibility to pancreatic cancer and its functional characterisation: the PANcreatic Disease ReseArch (PANDoRA) consortium. Digestive Liver Dis. 2013;45(2):95–99. doi:10.1016/j.dld.2012.09.014
5. Huang C, Xie K. Analysis of the potential for pancreatic cancer metastasis in vitro and in vivo. Methods Mol Biol. 2013;980:301–319.
6. Hinton J, Callan R, Bodine C, et al. Potential epigenetic biomarkers for the diagnosis and prognosis of pancreatic ductal adenocarcinomas. Expert Rev Mol Diagn. 2013;13(5):431–443. doi:10.1586/erm.13.38
7. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
8. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
9. Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67(1):129–139. doi:10.1007/s13105-010-0050-6
10. Lekka E, Hall J. Non-coding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900.
11. Sun LL, Li WD, Lei FR, Li XQ. The regulatory role of microRNAs in angiogenesis-related diseases. J Cell Mol Med. 2018;22:4568–4587. doi:10.1111/jcmm.13700
12. Dragomir M, Mafra ACP, Dias SMG, Vasilescu C, Calin GA. Using microRNA networks to understand cancer. Int J Mol Sci. 2018;19(7):1871. doi:10.3390/ijms19071871
13. Graner MW, Schnell S, Olin MR. Tumor-derived exosomes, microRNAs, and cancer immune suppression. Semin Immunopathol. 2018;40:505–515. doi:10.1007/s00281-018-0689-6
14. Toiyama Y, Okugawa Y, Fleshman J, Richard Boland C, Goel A. MicroRNAs as potential liquid biopsy biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta. 2018;1870(2):274–282.
15. Bryzgunova OE, Konoshenko MY, Laktionov PP. MicroRNA-guided gene expression in prostate cancer: literature and database overview. J Gene Med. 2018;20(5):e3016. doi:10.1002/jgm.v20.5
16. Drakaki A, Iliopoulos D. MicroRNA-gene signaling pathways in pancreatic cancer. Biomed J. 2013;36(5):200–208. doi:10.4103/2319-4170.119690
17. Guo S, Fesler A, Wang H, Ju J. microRNA based prognostic biomarkers in pancreatic cancer. Biomarker Res. 2018;6:18. doi:10.1186/s40364-018-0131-1
18. Huang J, Liu J, Chen-Xiao K, et al. Advance in microRNA as a potential biomarker for early detection of pancreatic cancer. Biomarker Res. 2016;4:20. doi:10.1186/s40364-016-0074-3
19. Tomar S, Plotnik JP, Haley J, et al. ETS1 induction by the microenvironment promotes ovarian cancer metastasis through focal adhesion kinase. Cancer Lett. 2018;414:190–204. doi:10.1016/j.canlet.2017.11.012
20. Sato Y, Kanno S, Oda N, et al. Properties of two VEGF receptors, Flt-1 and KDR, in signal transduction. Ann N Y Acad Sci. 2000;902:201–205. doi:10.1111/j.1749-6632.2000.tb06314.x.
21. Dittmer J. The biology of the Ets1 proto-oncogene. Mol Cancer. 2003;2:29. doi:10.1186/1476-4598-2-29
22. Dittmer J. The role of the transcription factor Ets1 in carcinoma. Semin Cancer Biol. 2015;35:20–38. doi:10.1016/j.semcancer.2015.09.010
23. Shaikhibrahim Z, Wernert N. ETS transcription factors and prostate cancer: the role of the family prototype ETS-1 (review). Int J Oncol. 2012;40(6):1748–1754. doi:10.3892/ijo.2012.1380
24. Xu S, Ge J, Zhang Z, Zhou W. MiR-129 inhibits cell proliferation and metastasis by targeting ETS1 via PI3K/AKT/mTOR pathway in prostate cancer. Biomed Pharmacother. 2017;96:634–641. doi:10.1016/j.biopha.2017.10.037
25. Hua S, Lei L, Deng L, et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene. 2018;37(12):1624–1636. doi:10.1038/s41388-017-0057-3
26. Yang Z, He J, Gao P, et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8(69):113558–113570. doi:10.18632/oncotarget.v8i69
27. Qiu HJ, Lu XH, Yang SS, Weng CY, Zhang EK, Chen FC. MiR-769 promoted cell proliferation in human melanoma by suppressing GSK3B expression. Biomed Pharmacother. 2016;82:117–123. doi:10.1016/j.biopha.2016.04.052
28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
29. Ito T, Nakayama T, Ito M, Naito S, Kanematsu T, Sekine I. Expression of the ets-1 proto-oncogene in human pancreatic carcinoma. Mod Pathol. 1998;11(2):209–215.
30. Lefter LP, Dima S, Sunamura M, et al. Transcriptional silencing of ETS-1 efficiently suppresses angiogenesis of pancreatic cancer. Cancer Gene Ther. 2009;16(2):137–148. doi:10.1038/cgt.2008.65
31. Khanna A, Mahalingam K, Chakrabarti D, Periyasamy G. Ets-1 expression and gemcitabine chemoresistance in pancreatic cancer cells. Cell Mol Biol Lett. 2011;16(1):101–113. doi:10.2478/s11658-010-0043-z
32. Li C, Wang Z, Chen Y, et al. Transcriptional silencing of ETS-1 abrogates epithelial-mesenchymal transition resulting in reduced motility of pancreatic cancer cells. Oncol Rep. 2015;33(2):559–565. doi:10.3892/or.2014.3613
33. Li Z, Lin P, Gao C, et al. Integrin beta6 acts as an unfavorable prognostic indicator and promotes cellular malignant behaviors via ERK-ETS1 pathway in pancreatic ductal adenocarcinoma (PDAC). Tumour Biol. 2016;37(4):5117–5131. doi:10.1007/s13277-015-4353-7
34. Slotwinski R, Lech G, Slotwinska SM. MicroRNAs in pancreatic cancer diagnosis and therapy. Cent-Eur j Immunol. 2018;43(3):314–324. doi:10.5114/ceji.2018.80051
35. Ebrahimi S, Hosseini M, Ghasemi F, et al. Circulating microRNAs as potential diagnostic, prognostic and therapeutic targets in pancreatic cancer. Curr Pharm Des. 2016;22(42):6444–6450. doi:10.2174/1381612822666160817095047
36. Diab M, Muqbil I, Mohammad RM, Azmi AS, Philip PA. The role of microRNAs in the diagnosis and treatment of pancreatic adenocarcinoma. J Clin Med. 2016;5(6):59. doi:10.3390/jcm5060059
37. Tokuhara K, Ogata Y, Nakagawa M, Shirouzu K. Ets-1 expression in vascular endothelial cells as an angiogenic and prognostic factor in colorectal carcinoma. Int Surg. 2003;88(1):25–33.
38. Furlan A, Vercamer C, Bouali F, et al. Ets-1 controls breast cancer cell balance between invasion and growth. Int J Cancer. 2014;135(10):2317–2328. doi:10.1002/ijc.28881
39. Zheng L, Qi T, Yang D, et al. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8(1):e55719. doi:10.1371/journal.pone.0055719
40. Calli AO, Sari A, Cakalagaoglu F, Altinboga AA, Oncel S. ETS-1 proto-oncogene as a key newcomer molecule to predict invasiveness in laryngeal carcinoma. Pathol Res Pract. 2011;207(10):628–633. doi:10.1016/j.prp.2011.07.010
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.