Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-761 targets FGFR1 to suppress the malignancy of osteosarcoma by deactivating PI3K/Akt pathway
Authors Lv Z, Ma J, Wang J, Lu J
Received 11 March 2019
Accepted for publication 15 August 2019
Published 15 October 2019 Volume 2019:12 Pages 8501—8513
DOI https://doi.org/10.2147/OTT.S208263
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
This paper has been retracted.
Zhongzhe Lv, 1 Jinming Ma, 2 Jianchuan Wang, 1 Jianmin Lu 1
1Department of Orthopedics, Affiliated Zhongshan Hospital of Dalian University, Liaoning 116001, People’s Republic of China; 2School of Graduate Studies, Zunyi Medical University, Guizhou 563000, People’s Republic of China
Correspondence: Jianmin Lu
Department of Orthopedics, Affiliated Zhongshan Hospital of Dalian University, No. 6 Jiefang Road, Zhongshan District, Liaoning 116001, People’s Republic of China
Tel +86 181 467 5683
Email [email protected]
Purpose: MicroRNA-761 (miR-761) has been reported to be deregulated in many types of human cancers and play important roles in cancer genesis and progression. However, the biological roles of miR-761 in osteosarcoma (OS) and the underlying mechanisms remain largely unknown.
Methods: The expression of miR-761 in OS tissues and cell lines was analyzed using RT-qPCR. A series of gain-of-function tests were performed, and status of malignancy was evaluated on basis of proliferation, migration, invasion, and apoptosis using different assays to determine the regulatory roles of miR-761 in OS cells in vivo and in vitro. Notably, the mechanisms underlying the action of miR-761 in the pathogenesis of OS were investigated using bioinformatic analysis, luciferase reporter assay, RT-qPCR and Western blotting.
Results: The results showed that miR-761 expression was decreased in OS tissues and cell lines and is closely correlated with clinical stage and distant metastasis in OS patients. Patients with OS having low miR-761 expression showed worse prognosis compared to OS patients with high miR-761 expression. Restoring the miR-761 expression level decreased OS cell proliferation, migration, and invasion in vitro; promoted cell apoptosis in vitro; and impaired tumor growth in vivo. In addition, fibroblast growth factor receptor 1 (FGFR1) was found as a direct target gene of miR-761 in OS cells. Furthermore, silencing FGFR1 expression stimulated the tumor-suppressing roles of miR-761 upregulation in OS cells, whereas the activity of miR-761 overexpression in OS cells was abolished by the restoration of FGFR1 expression. Moreover, restoration of miR-761 expression deactivated the PI3K/Akt pathway in vitro and in vivo.
Conclusion: These results suggest that miR-761 plays anti-cancer roles in OS by directly targeting FGFR1 and deactivating the PI3K/Akt pathway. The newly identified miR-761/FGFR1/PI3K/Akt pathway partially illustrates the mechanism of OS pathogenesis and presents a novel candidate therapeutic target for antitumor therapy.
Keywords: microRNA-761, osteosarcoma, fibroblast growth factor receptor 1, PI3K/Akt pathway, pathogenesis
Introduction
Osteosarcoma (OS), which derives from bone-forming mesenchymal cells, remains the most prevailing aggressive malignant bone tumor.1 It accounts for approximately 5% of all the pediatric tumors and is typically diagnosed in children and young adults between the ages of 10 and 20 years.2 OS normally develops in the proximal end of the long bones and spine and is characterized by high recurrence rate, high malignant potential, and frequent relapses.3,4 The combination treatment involves tumor excision, neoadjuvant therapy, and adjuvant chemotherapy which has notably improved the clinical outcomes and long-term survival of the patients in the last several decades.5 However, about one-third of the OS patients with metastases or recurrence have been observed to have a dismal prognosis.6 Several reports have demonstrated that alterations in the genome and transcriptome contribute towards the pathogenesis of OS;7–9 unfortunately, many of the putative mechanisms which affect the genesis and development of OS remain to be investigated. Thus, an intensive investigation is crucially needed to study the molecular mechanisms responsible for the formation and progression of OS and its significance to find novel therapeutic targets for anticancer therapy.
MicroRNAs (miRNAs) are a large family of endogenous, non-coding, and short RNA molecules comprising 19–25 nucleotides in length.10 miRNAs have been considered as vital gene regulators. They base-pair with complementary sequences in the 3′-untranslated regions (3′-UTRs) of their target genes causing translational suppression and mRNA cleavage.11 Until now, more than 1,000 miRNAs have been validated in the human genome, and these miRNAs are expected to negatively modulate about 60% of all human protein-coding genes.12 Emerging studies have indicated that miRNAs are abnormally expressed in various human diseases including malignant tumor.13–16 The dysregulated miRNAs may contribute to the malignant progression of OS and participate in the regulation of many cellular biological processes such as cell proliferation, cell cycle, apoptosis, invasion, and metastasis.17–19 Consequently, miRNAs can be fascinating therapeutic targets for the treatment of OS patients, and thus, it is imperative to further elucidate the biological functions of miRNAs in the pathogenesis and progression of OS.
Recently, miR-761 has been reported to be downregulated in many types of human cancer, and has been revealed to play important roles in the genesis and progression of cancer.20–25 However, the biological roles of miR-761 in OS and its underlying mechanism remains largely unknown. In the present study, we detected miR-761 expression in OS and investigated the regulatory effects of miR-761 overexpression on OS progression. In addition, the mechanisms underlying the functional roles of miR-761 in OS pathogenesis were analyzed. Our research provides a novel theoretical basis for the diagnosis and treatment of OS.
Materials and methods
Human tissue specimens
A total of 61 pairs of OS tissues and the corresponding adjacent normal tissues were collected from patients with OS that were admitted to Affiliated Zhongshan Hospital of Dalian University. Tissues were surgically removed, flash frozen in liquid nitrogen, and stored at −80 °C until further use. All the patients enrolled in the study neither received radiotherapy, chemotherapy, nor any other treatments prior to surgery. This study was approved by the Ethics Committee of Affiliated Zhongshan Hospital of Dalian University, and written informed consent was obtained from all patients before their enrollment.
Cell culture
Normal human osteoblast cells (hFOB1.19) and four human OS cell lines (U2OS, MG-63, HOS and SAOS-2) were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 1% of penicillin/streptomycin was used to culture the cells (Invitrogen, Carlsbad, CA, USA). The cells were incubated at 37 °C in a humidified atmosphere supplied with 5% CO2. Cells in logarithmic growth phase were used for subsequent functional analysis.
Transfection assay
Agomir-761, agomir-NC, small interfering RNA (siRNA) targeting Fibroblast growth factor receptor 1 (FGFR1) [si-FGFR1], and scrambled control siRNA (si-ctrl) were obtained from Shanghai GenePharma Co Ltd (Shanghai, China). To restore FGFR1 expression, FGFR1 overexpression plasmid (pCMV-FGFR1) and an empty pCMV plasmid were custom synthesized from Integrated Biotech Solutions (Shanghai, China). Transient transfection was conducted using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) diluted in Opti-MEM (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue samples or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Concentration and quality of the extracted RNA was determined using Nanodrop® ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc, Pittsburgh, PA, USA). To quantify miR-761 expression, total RNA was reversed transcribed into complementary DNA (cDNA) using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). Further, qPCR was performed using TaqMan miRNA PCR Kit (Applied Biosystems, Carlsbad, CA, USA). To evaluate FGFR1 mRNA expression, cDNA was synthesized from total RNA using PrimeScript™ RT Reagent kit followed by qPCR with SYBR® Premix Ex Taq™ II Kit (both from Takara Bio Inc, Otsu, Japan). miR-761 and FGFR1 mRNA expression were standardized to the expression levels of U6 snRNA and GAPDH, respectively. Relative gene expression was analyzed by the 2−ΔΔCq method.26
Cell counting kit-8 (CCK-8) assay
Transfected cells were incubated at 37 °C supplied with 5% CO2 for 24 h, then harvested, and re-suspended in DMEM containing 10% FBS. Transfected cells were seeded in 96-well plates (3000 cells/well). Cell proliferation was determined by CCK-8 assay at different time points: day 0, day 1, day 2, and day 3 after seeding. Overall, 10 μL of CCK-8 solution (Dojindo, Tokyo, Japan) was added into each well and then incubated at 37 °C with 5% CO2 for additional 2 h. The absorbance value was determined at 450 nm using an ELISA microplate reader (Bio-Rad Laboratories Inc, Hercules, CA, USA).
Cell apoptosis assay
Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Biolegend, San Diego, CA, USA) was used to determine cellular apoptosis. Transfected cells were collected, washed thrice with ice-cold phosphate buffered saline (PBS), and then resuspended in 100 µL of binding buffer. The cells were stained with 5 µL Annexin V-FITC and 5 µL propidium iodide in the dark for 30 min and then subjected to flow cytometry (FACScan™; BD Biosciences, Franklin Lakes, NJ, USA) for detecting cell apoptosis.
Transwell migration and invasion assay
Transwell chambers (Corning, Incorporated, Corning, NY, USA) were pre-coated with Matrigel (BD Bioscience, San Jose, CA, USA) and used to evaluate the invasion ability of the cultured cells. Transfected cells were harvested after 48 h of incubation and resuspended in serum-free DMEM. A total of 200 μL serum-free DMEM with 5× 105 cells was seeded into the upper chamber inserts while the bottom chambers were added with 600 μL DMEM containing 10% FBS. Following 24 h incubation, the non-invaded cells of the upper chamber insert were carefully wiped using a cotton swab. The invaded cells were fixed with 95% methanol, stained with 0.1% crystal violet, and visualized under a microscope. The number of invaded cells was counted from five randomized visual fields for each chamber under a microscope (IX53, Olympus Corporation, Tokyo, Japan). Similarly, transwell migration assay was performed using the transwell chambers which were not coated with Matrigel.
Tumor xenograft assay
All animal model experiments were approved by the Laboratory Animal Ethics Committee of Affiliated Zhongshan Hospital of Dalian University and followed the guidance of Animal Protection Law of the People’s Republic of China-2009. 6-week-old BALB/c nude mice were bought from the Shanghai Laboratory Animal Center (Chinese Academy of Sciences, Shanghai, China) and kept under specific pathogen-free conditions. Cells transfected with agomir-761 or agomir-NC were harvested after 24 h of incubation and injected subcutaneously into the flank of nude mice. The width and length of the tumor xenograft was recorded every four days and tumor volumes were calculated using the formula: 1/2×tumor length×tumor width.2 All nude mice were euthanized by cervical dislocation at four weeks after injection. Tumor xenograft was excised and stored until further use.
Bioinformatic analysis and luciferase reporter assay
The putative targets of miR-761 were predicted using TargetScan v7.1 (www.targetscan.org) and microRNA.org (www.microrna.org) database.
The human FGFR1 3′-UTR region containing the wild-type (WT) or mutant (MUT) miR-761 binding site was amplified by Shanghai GenePharma Co Ltd, cloned into a pmirGLO luciferase reporter gene (Promega, Madison, WI, USA) and referred as pmirGLO-WT-FGFR1-3′-UTR and pmirGLO-MUT-FGFR1-3′-UTR, respectively. For reporter assay, cells were seeded in a 24-well plate and co-transfected with either pmirGLO-WT-FGFR1-3′-UTR or pmirGLO-MUT-FGFR1-3′-UTR, and treated with agomir-761 or agomir-NC using Lipofectamine 2000. Luciferase activities were measured using dual-luciferase reporter assay (Promega, Madison, WI, USA) at 48 h post-transfection following the manufacturer’s instruction. Firefly luciferase activity was standardized to Renilla luciferase activity.
Western blot analysis
Transfected cells were lysed in radioimmunoprecipitation Assay buffer (Beyotime Institute of Biotechnology, Haimen, China) after 72 h of incubation. The concentration of total protein was determined using a BCA Protein Assay kit (Beyotime Institute of Biotechnology, Haimen, China). Equivalent proteins were separated by SDS-PAGE (10% polyacrylamide gel) and then transferred onto polyvinylidene difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked at room temperature for 2 h with Tris-buffered saline with Tween-20 (TBST) containing 5% dried skimmed milk. Subsequently, the membranes were incubated with primary antibodies overnight at 4 °C. Following three washes with TBST, protein signals were detected using horseradish peroxidase-conjugated secondary antibody (ab6734 and ab205719; Santa Cruz Biotechnology, CA, USA) for 2 h at room temperature and developed using Pierce™ ECL Western Blotting Substrate (Pierce Biotechnology Inc, Rockford, IL, USA). The primary antibodies used in this study were as follows: mouse anti-human monoclonal FGFR1 antibody (ab824; Abcam; Cambridge, MA, USA), rabbit anti-human polyclonal p-PI3K antibody (ab182651; Abcam; Cambridge, MA, USA), mouse anti-human monoclonal pi3k (ab189403; Abcam; Cambridge, MA, USA), mouse anti-human monoclonal p-Akt antibody (sc-81433; Santa Cruz Biotechnology, CA, USA), mouse anti-human monoclonal Akt antibody (sc-56878; Santa Cruz Biotechnology, CA, USA), and mouse anti-human monoclonal GAPDH antibody (sc-47724; Santa Cruz Biotechnology, CA, USA).
Statistical analysis
All data were represented as the mean ± standard deviation (SD). The clinical association analysis was determined with Chi-square test. One-way analysis of variance followed by a Student-Newman-Keuls post-hoc test was used to compare the significant differences between more than two groups. The significant difference between the two groups was analyzed using Student’s t-test. Spearman’s correlation analysis was used to examine the putative expression correlations between miR-761 and FGFR1 mRNA levels in OS tissues. Overall survival was analyzed using the Kaplan-Meier method and the significant difference between overall survival was examined by log-rank test. All statistical analysis were evaluated with SPSS version 18 (SPSS Inc, Chicago, IL, USA). P < 0.05 was considered statistically significant.
Results
Mir-761 is remarkably downregulated in OS
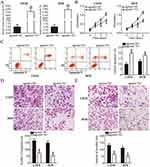
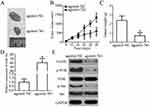
To evaluate the role of miR-761 in the genesis and progression of OS, we first detected miR-761 expression level in the 61 pairs of OS tissues and corresponding adjacent normal tissues. The RT-qPCR analysis revealed that miR-761 was significantly downregulated in OS tissues compared to its expression in adjacent normal tissues (Figure 1A, P<0.05). Further, RT-qPCR analysis of in vitro cancer cell lines showed that the expression level of miR-761 was lower in OS cell lines (U2OS, MG-63, HOS, and SAOS-2) compared to its expression in normal human osteoblast cells (hFOB 1.19) (Figure 1B, P<0.05), thereby supporting the data obtained from OS tissues.
To confirm its clinical significance, miRNA data of all OS patients was divided into miR-761-low or miR-761-high expression groups with the median value as cut-off. A low miR-761 expression was positively correlated with the clinical stage (P=0.021) and distant metastasis (P=0.002) in patients with OS (Table 1). The prognostic significance of miR-761 in patients with OS was also studied where the results indicated that miR-761-low group showed an obvious shorter overall survival compared to patients in miR-761-high group (Figure 1C, P=0.0213). These results implied that miR-761 expression was decreased in OS which may be closely associated with the tumor development.
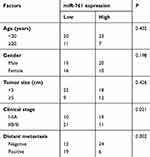 |
Table 1 The relationship between miR-761 and clinicopathological features in patients with OS |
miR-761 attenuates OS cell proliferation, migration, and invasion and promotes cell apoptosis in vitro
To illustrate the functional significance of miR-761 in OS progression, U2OS and HOS cell lines were selected for functional assays. The two cell lines were transfected with agomir-761 or agomir-NC. As shown in Figure 2A, RT-qPCR analysis revealed that miR-761 was significantly overexpressed in U2OS and HOS cells after transfection with agomir-761 (P<0.05). The functional role of miR-761 overexpression in OS cell proliferation and apoptosis was determined using CCK-8 assay and flow cytometry analysis, respectively. The proliferative ability of both the cell lines (U2OS and HOS) was evidently decreased when miR-761 was upregulated (Figure 2B, P<0.05). Additionally, the ratio of apoptotic cells was found to be increased in U2OS and HOS cells after gain of miR-761 (Figure 2C, P<0.05). Furthermore, transwell migration and invasion assays were used to determine the significance of miR-761 overexpression on the migration and invasion property of the OS cells in vitro. It was observed that transfection with agomir-761 noticeably suppressed the metastatic ability of U2OS and HOS cells compared to the metastatic capability of cells transfected with agomir-NC (Figure 2D and E, P<0.05). These results showed that miR-761 may play a tumor suppressive role in OS progression.
FGFR1 is a direct target gene of miR-761 in OS cells
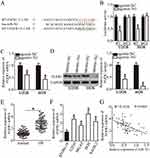
To reveal the mechanisms underlying the suppressive effects of miR-761 in OS cells, the putative targets of miR-761 were analyzed using TargetScan v7.1 and microRNA.org. The bioinformatic analysis predicted that the 3′-UTR of FGFR1 has a potential miR-761 binding site (Figure 3A). To show a direct relationship between miR-761 and the 3′-UTR of FGFR1, luciferase reporter plasmids were constructed and co-transfected with agomir-761 or agomir-NC in U2OS and HOS cells. Luciferase reporter assay was performed to detect the luciferase activity which revealed that miR-761 overexpression attenuated the luciferase activity of the plasmid expressing wild-type 3′-UTR FGFR1 in U2OS and HOS cells (Figure 3B, P<0.05). However, when the binding sequences were mutated, there was no inhibitory activity observed.
To investigate whether the endogenous FGFR1 was affected by miR-761, FGFR1 expression was detected in the agomir-761 or agomir-NC transfected U2OS and HOS cells. The FGFR1 mRNA (Figure 3C, P<0.05) as well as protein (Figure 3D, P<0.05) levels were significantly decreased in miR-761 overexpressing-U2OS and HOS cells, as shown by RT-qPCR and Western blot analysis. Furthermore, the expression level of FGFR1 mRNA was significantly higher in OS tissues compared to their mRNA levels in adjacent normal tissues (Figure 3E, P<0.05). The expression of FGFR1 in OS cell lines and normal human osteoblast cells hFOB 1.19 was also detected through RT-qPCR analysis. The mRNA level of FGFR1 was notably increased in all four OS cell lines than in hFOB 1.19 (Figure 3F, P<0.05). Additionally, an inverse relation between miR-761 and FGFR1 mRNA levels in the OS tissues was revealed using Spearman’s correlation analysis (Figure 3G; R2=0.3226, P<0.0001). Collectively, we observed that miR-761 suppressed the expression of FGFR1 by binding directly to its 3′-UTR. Thus, FGFR1 was shown to be a direct target gene of miR-761 in OS cells.
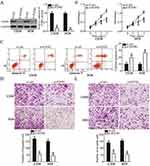
Silencing FGFR1 imitates the tumor-suppressing roles of miR-761 in OS cells
As FGFR1 was identified as a direct target of miR-761 in OS cells, we performed loss-of-function assays to analyze the role of FGFR1 in OS progression. U2OS and HOS cells were transfected with si-FGFR1 or si-ctrl and Western blot analysis was used to determine the transfection efficiency. We observed that the transfection of si-FGFR1 effectively inhibited the expression of FGFR1 in U2OS and HOS cells (Figure 4A, P<0.05). FGFR1 deficiency significantly decreased proliferation (Figure 4B, P<0.05) and increased apoptosis (Figure 4C, P<0.05) in U2OS and HOS cells which was revealed by CCK-8 assay and flow cytometry analysis. Furthermore, transwell migration and invasion assay revealed that the silenced FGFR1 expression remarkably impaired the migratory (Figure 4D, P<0.05) and invasive (Figure 4E, P<0.05) capabilities of U2OS and HOS cells. Overall, these results indicated that the functions of FGFR1 silencing in OS cells were similar to the functions stimulated by miR-761-mediated downregulation of FGFR1. Thus, FGFR1 is a downstream target of miR-761 in OS cells.
Tumor-suppressing roles of miR-761 in OS cells were mediated by FGFR1
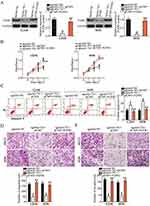
With FGFR1 confirmed as a direct target of miR-761 from the above experiments, we further investigated whether FGFR1 mediated the inhibitory effects of miR-761 upregulation in OS cells. We transfected the U2OS and HOS cells with agomir-761 and pCMV or pCMV-FGFR1. Western blot analysis revealed that the FGFR1 protein expression was decreased on miR-761 overexpression which could be restored in U2OS and HOS cells by pCMV-FGFR1 co-transfection (Figure 5A, P<0.05). Consistently, functional analysis confirmed that the proliferation (Figure 5B, P<0.05), apoptosis (Figure 5C, P<0.05), migration (Figure 5D, P<0.05) and invasion (Figure 5E, P<0.05) capabilities of U2OS and HOS cells were affected by miR-761 overexpression. However, co-transfection with pCMV-FGFR1abolished the effects induced by miR-761 upregulation. Thus, these findings showed that the tumor suppressor activity of miR-761 in OS cells was partially mediated by FGFR1.
Upregulation of miR-761 inhibits the activation of PI3K/Akt pathway in OS cells through negatively regulating FGFR1 expression
Previous studies had reported that the PI3K/Akt pathway was activated by FGFR1 in OS cells.27,28 Accordingly, whether the PI3K/Akt pathway could be negatively regulated by miR-761 in OS cells was investigated in this study. pCMV-FGFR1 or pCMV was co-transfected with agomir-761 in U2OS and HOS cells. Western blotting was used to evaluate the expression levels of important signaling molecules in PI3K/Akt pathway. Restoring miR-761 expression notably reduced the protein levels of p-PI3K and p-Akt in U2OS and HOS cells, and this outcome could be abolished by co-transfection with pCMV-FGFR1 (Figure 6). These results suggested that miR-761 suppressed the activation of PI3K/Akt pathway in OS cells by decreasing FGFR1 expression.
miR-761 targets FGFR1 to inhibit the tumor growth of OS in vivo
To determine the influence of miR-761 on tumor growth, we performed tumor xenograft assay by injecting agomir-761-transfected or agomir-NC-transfected HOS cells in nude mice. The results showed that the tumor growth (Figure 7A and B, P<0.05) and weight (Figure 7C, P<0.05) of the xenograft in the agomir-761 group was notably smaller compared to that in the agomir-NC group. Total RNA from tumor xenografts was isolated and RT-qPCR analysis was performed. It revealed that the expression of miR-761 was upregulated in the tumor xenografts isolated from agomir-761-transfected HOS cells (Figure 7D, P<0.05). Western blot analysis showed that FGFR1, p-PI3K, and p-Akt protein expression was decreased in the agomir-761 group compared to their protein expressions in the agomir-NC group (Figure 7E). Thus, the results showed that restoring miR-761 expression suppressed the OS tumor growth in vivo by reducing FGFR1 expression and inactivating the PI3K/Akt pathway.
Discussion
Aberration in miRNA expression has been consistently reported in many types of human cancer including OS.29–31 miRNAs may play either oncogenic or tumor suppressive roles and contribute in the regulation of malignant behavior of OS cells.32–34 Strategies for prohibiting different types of human malignancy using miRNAs have been proposed in several literatures.35–37 Thus, the functional roles of miRNAs in the growth mechanism of OS should be investigated which might be useful in finding novel therapeutic strategies for treating patients with OS. We are the first to detect the expression of miR-761 in OS and analyze its clinical significance in patients with OS. Additionally, we investigated the comprehensive roles of miR-761 in OS progression using a series of functional experiments. Notably, the mechanisms responsible for the tumor-suppressing action on miR-761 upregulation in OS cells were also thoroughly analyzed in this study.
miR-761 is downregulated in colorectal20–22 and ovarian23 cancers. The downregulation of miR-761 is related with the tumor size, tumor grade, and tumor stage in patients with colorectal cancer.20,21 Patients with colorectal cancer having low miR-761 expression show worse 5-year overall survival compared to patients with high miR-761 expression.20 On the contrary, miR-761 is upregulated in non-small cell lung cancer,24,38 breast cancer25 and hepatocellular carcinoma.39 However, there has been no study to investigate the expression level and clinical significance of miR-761 in OS. In this study, we found that miR-761 expression was decreased in OS tissues as well as cell lines. Investigation of the relation between miR-761 expression and clinicopathological features revealed that a low miR-761 expression was correlated to clinical stage and distant metastasis. The overall survival of patients with OS expressing low miR-761 levels was shorter compared to OS patients with high miR-761 expression. These observations suggest that miR-761 is a potential biomarker for the diagnosis and prognosis of patients with OS.
miR-761 is characterized as a tumor-suppressing miRNA in many types of human cancer. For instance, restoring miR-761 expression decreases cell proliferation, cell cycle, colony formation, migration, and invasion in colorectal cancer.20–22 Moreover, it has been reported that upregulation of miR-761 increases the chemosensitivity of colorectal cancer cells to 5-Fluorouracil.21 Shi and Zhang have reported that restoring miR-761 expression attenuated the proliferation and invasion of ovarian cancer cells.23 On the contrary, miR-761 plays oncogenic roles in non-small cell lung cancer by promoting cell growth and metastasis in vitro.24 In triple-negative breast cancer25 and hepatocellular carcinoma,39 miR-761 overexpression enhances cell growth and metastasis in vitro as well as in vivo. However, the detailed roles of miR-761 in OS have not been reported yet. Here, we showed that the overexpressed miR-761 inhibits the proliferation, migration, and invasion of OS cells in vitro and impaired OS tumor growth in vivo. Further, we also proved that miR-761 overexpression promotes apoptosis in OS cells. These results suggest that miR-761 can be an effective target for the therapy of patients with OS.
In previous studies, several genes such as Rab3D,20 FOXM1,21 HDAC1,22 MSI1,23 ING4,24 TIMP2,24 TRIM29,25 and mitofusin-239 have been reported to be direct targets of miR-761. In our current study, FGFR1 was confirmed as a direct target gene of miR-761 and its deficiency was essential for the functional activity of miR-761 in OS cells. FGFR1 expression was found to be increased in OS and high FGFR1 expression was significantly correlated with the location of the malignant lesion, advanced clinical stage, and distant metastasis.28 In addition, OS patients with high FGFR1 expression have shown poor response to neo-adjuvant chemotherapy compared to OS patients with low FGFR1 expression.40 It has been reported that FGFR1 upregulation contributes to the malignancy of OS by regulating a number of oncogenic properties such as cell viability, proliferation, apoptosis, metastasis, and epithelial-mesenchymal transition.40–43 The present study revealed that miR-761 directly targets FGFR1 in OS cells and deactivates the PI3K/Akt pathway cells in vitro as well as in vivo, thereby suppressing OS progression. Thus, silencing FGFR1 using miR-761-mediated targeted therapy can be a promising therapeutic strategy for patients with OS. FGFR1 was also implicated in the regulation of RAS/ERK1/2 pathway,44 Src/NF-κB pathway,45 and Wnt pathway.46 However, in this study, we did not explored whether miR-761 was implicated in the regulation of these pathways. It was a limitation and we will resolve it in the near future.
Conclusion
In conclusion, miR-761 is downregulated in OS and its low expression predicts poor overall survival of patients with OS. Upregulation of miR-761 prohibits OS progression in vitro and in vivo by directly targeting FGFR1 via deactivating the PI3K/Akt pathway. A better understanding of the miR-761-FGFR1-PI3K/Akt pathway in OS may further elucidate the mechanism underlying OS progression and provide a potential target for the anticancer therapy in OS patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Poletajew S, Fus L, Wasiutynski A. Current concepts on pathogenesis and biology of metastatic osteosarcoma tumors. Ortop Traumatol Rehabil. 2011;13(6):537–545.
2. Ando K, Mori K, Verrecchia F, Marc B, Redini F, Heymann D. Molecular alterations associated with osteosarcoma development. Sarcoma. 2012;2012:523432. doi:10.1155/2012/523432
3. Pakos EE, Nearchou AD, Grimer RJ, et al. Prognostic factors and outcomes for osteosarcoma: an international collaboration. Eur J Cancer. 2009;45(13):2367–2375. doi:10.1016/j.ejca.2009.03.005
4. Katonis P, Datsis G, Karantanas A, et al. Spinal osteosarcoma. Clin Med Insights Oncol. 2013;7:199–208. doi:10.4137/CMO.S10099
5. Subbiah V, Kurzrock R. Phase 1 clinical trials for sarcomas: the cutting edge. Curr Opin Oncol. 2011;23(4):352–360. doi:10.1097/CCO.0b013e3283477a94
6. Angelini A, Ceci F, Castellucci P, et al. The role of (18)F-FDG PET/CT in the detection of osteosarcoma recurrence. Eur J Nucl Med Mol Imaging. 2017;44(10):1712–1720. doi:10.1007/s00259-017-3698-0
7. Bielack SS, Kempf-Bielack B, Delling G, et al. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol. 2002;20(3):776–790. doi:10.1200/JCO.2002.20.3.776
8. Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33(27):3029–3035. doi:10.1200/JCO.2014.59.4895
9. Shaikh AB, Li F, Li M, et al. Present advances and future perspectives of molecular targeted therapy for osteosarcoma. Int J Mol Sci. 2016;17(4):506. doi:10.3390/ijms17040506
10. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
11. Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35(3):222–229. doi:10.1038/nbt.3802
12. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi:10.1101/gr.082701.108
13. Piasecka D, Braun M, Kordek R, Sadej R, Romanska H. MicroRNAs in regulation of triple-negative breast cancer progression. J Cancer Res Clin Oncol. 2018. doi:10.1007/s00432-018-2689-2
14. Mukai N, Nakayama Y, Murakami S, et al. Potential contribution of erythrocyte microRNA to secondary erythrocytosis and thrombocytopenia in congenital heart disease. Pediatric research. 2018;83(4):866–873.
15. Martinez B, Peplow PV. MicroRNAs in parkinson’s disease and emerging therapeutic targets. Neural Regener Res. 2017;12(12):1945–1959. doi:10.4103/1673-5374.221147
16. Wang Y, Cheng Q, Liu J, Dong M. Leukemia stem cell-released microvesicles promote the survival and migration of myeloid leukemia cells and these effects can be inhibited by MicroRNA34a overexpression. Stem Cells Int. 2016;2016:9313425. doi:10.1155/2016/1243659
17. Rongxin S, Pengfei L, Li S, Xiaochen J, Yihe H. MicroRNA-340-5p suppresses osteosarcoma development by down-regulating the Wnt/beta-catenin signaling pathway via targeting the STAT3 gene. Eur Rev Med Pharmacol Sci. 2019;23(3):982–991. doi:10.26355/eurrev_201902_16985
18. Xie W, Xiao J, Wang T, Zhang D, Li Z. MicroRNA-876-5p inhibits cell proliferation, migration and invasion by targeting c-Met in osteosarcoma. J Cell Mol Med. 2019. doi:10.1111/jcmm.14217
19. Zhang Z, Luo G, Yu C, Yu G, Jiang R, Shi X. MicroRNA-493-5p inhibits proliferation and metastasis of osteosarcoma cells by targeting kruppel-like factor 5. J Cell Physiol. 2019;234(8):13525–13533.
20. Ren Y, Shi G, Jiang P, Meng Q. MicroRNA-761 is downregulated in colorectal cancer and regulates tumor progression by targeting Rab3D. Exp Ther Med. 2019;17(3):1841–1846. doi:10.3892/etm.2018.7126
21. Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2018;9(1):321–331. doi:10.18632/oncotarget.20109
22. Xiong W, Yang S, Zhang W, Chen Y, Wang F. MiR-761 inhibits colorectal cancer cell proliferation and invasion through targeting HDAC1. Pharmazie. 2019;74(2):111–114. doi:10.1691/ph.2019.8756
23. Shi C, Zhang Z. miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma. Tumour Biol. 2016;37(4):5437–5443. doi:10.1007/s13277-015-4377-z
24. Yan A, Yang C, Chen Z, Li C, Cai L. MiR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 2015;37(1):55–66. doi:10.1159/000430333
25. Guo GC, Wang JX, Han ML, Zhang LP, Li L. microRNA-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression. Cell Oncol. 2017;40(2):157–166. doi:10.1007/s13402-016-0312-6
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
27. Kamura S, Matsumoto Y, Fukushi JI, et al. Basic fibroblast growth factor in the bone microenvironment enhances cell motility and invasion of Ewing’s sarcoma family of tumours by activating the FGFR1-PI3K-Rac1 pathway. Br J Cancer. 2010;103(3):370–381. doi:10.1038/sj.bjc.6605775
28. Gao G, Tian Z, Zhu HY, Ouyang XY. miRNA-133b targets FGFR1 and presents multiple tumor suppressor activities in osteosarcoma. Cancer Cell Int. 2018;18:210. doi:10.1186/s12935-018-0696-7
29. Liu W, Zhou Z, Zhang Q, et al. Overexpression of miR-1258 inhibits cell proliferation by targeting AKT3 in osteosarcoma. Biochem Biophys Res Commun. 2019;510(3):479–486. doi:10.1016/j.bbrc.2019.01.139
30. Ren J, Yang M, Xu F, Chen J. microRNA-758 inhibits the malignant phenotype of osteosarcoma cells by directly targeting HMGA1 and deactivating the Wnt/beta-catenin pathway. Am J Cancer Res. 2019;9(1):36–52.
31. Li L, Wang X, Liu D. MicroRNA-185 inhibits proliferation, migration and invasion in human osteosarcoma MG63 cells by targeting vesicle-associated membrane protein 2. Gene. 2019;696:80–87. doi:10.1016/j.gene.2019.01.034
32. Shekhar R, Priyanka P, Kumar P, et al. The microRNAs miR-449a and miR-424 suppress osteosarcoma by targeting cyclin A2 expression. J Biol Chem. 2019. doi:10.1074/jbc.RA118.005778
33. Zhan FB, Zhang XW, Feng SL, et al. MicroRNA-206 reduces osteosarcoma cell malignancy in vitro by targeting the PAX3-MET axis. Yonsei Med J. 2019;60(2):163–173. doi:10.3349/ymj.2019.60.2.163
34. Ma G, Zhang C, Luo W, Zhao JL, Wang X, Qian Y. Construction of microRNA-messenger networks for human osteosarcoma. J Cell Physiol. 2019. doi:10.1002/jcp.28107
35. Hosseinahli N, Aghapour M, Duijf PHG, Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol. 2018;233(8):5574–5588. doi:10.1002/jcp.26514
36. Yang G, Zhang W, Yu C, Ren J, An Z. MicroRNA let-7: regulation, single nucleotide polymorphism, and therapy in lung cancer. J Cancer Res Ther. 2015;11(Suppl 1):C1–C6. doi:10.4103/0973-1482.163830
37. Li Z, Yu X, Shen J, Law PT, Chan MT, Wu WK. MicroRNA expression and its implications for diagnosis and therapy of gallbladder cancer. Oncotarget. 2015;6(16):13914–13921. doi:10.18632/oncotarget.4227
38. Zhang MY, Zhang ZL, Cui HX, Wang RK, Fu L. Long non-coding RNA FENDRR inhibits NSCLC cell growth and aggressiveness by sponging miR-761. Eur Rev Med Pharmacol Sci. 2018;22(23):8324–8332. doi:10.26355/eurrev_201812_16530
39. Zhou X, Zhang L, Zheng B, et al. MicroRNA-761 is upregulated in hepatocellular carcinoma and regulates tumorigenesis by targeting mitofusin-2. Cancer Sci. 2016;107(4):424–432. doi:10.1111/cas.12904
40. Fernanda Amary M, Ye H, Berisha F, et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer Med. 2014;3(4):980–987. doi:10.1002/cam4.268
41. Zhou W, Zhu Y, Chen S, Xu R, Wang K. Fibroblast growth factor receptor 1 promotes MG63 cell proliferation and is associated with increased expression of cyclin-dependent kinase 1 in osteosarcoma. Mol Med Rep. 2016;13(1):713–719. doi:10.3892/mmr.2015.4597
42. Weekes D, Kashima TG, Zandueta C, et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35(22):2948. doi:10.1038/onc.2015.420
43. Ling L, Tan SK, TH G, et al. Targeting the heparin-binding domain of fibroblast growth factor receptor 1 as a potential cancer therapy. Mol Cancer. 2015;14:136. doi:10.1186/s12943-014-0278-9
44. Palumbo P, Petracca A, Maggi R, et al. A novel dominant-negative FGFR1 variant causes hartsfield syndrome by deregulating RAS/ERK1/2 pathway. Eur J Hum Genet. 2019;27(7):1113–1120. doi:10.1038/s41431-019-0350-4
45. Lai SW, Bamodu OA, Tsai WC, et al. The therapeutic targeting of the FGFR1/Src/NF-kappaB signaling axis inhibits pancreatic ductal adenocarcinoma stemness and oncogenicity. Clin Exp Metastasis. 2018;35(7):663–677. doi:10.1007/s10585-018-9919-5
46. Nguyen TM, Kabotyanski EB, Dou Y, et al. FGFR1-activated translation of WNT pathway components with structured 5ʹ UTRs is vulnerable to inhibition of EIF4A-dependent translation initiation. Cancer Res. 2018;78(15):4229–4240. doi:10.1158/0008-5472.CAN-18-0631
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.