Back to Journals » OncoTargets and Therapy » Volume 12
MicroRNA-675 directly targets MAPK1 to suppress the oncogenicity of papillary thyroid cancer and is sponged by long non-coding RNA RMRP
Authors Wang J, Xiao T, Zhao M
Received 25 April 2019
Accepted for publication 1 August 2019
Published 6 September 2019 Volume 2019:12 Pages 7307—7321
DOI https://doi.org/10.2147/OTT.S213371
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
This paper has been retracted.
Junyi Wang,* Tiantian Xiao,* Ming Zhao
Department of Endocrinology, Geriatric Research Center, JinLing Hospital, Nanjing, Medical School of Nanjing University, Jiangsu 210002, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ming Zhao
Department of Endocrinology, Geriatric Research Center, JinLing Hospital, Medical School of Nanjing University, No. 305 Zhongshan East Road, Nanjing, Jiangsu 210002, People’s Republic of China
Tel +86 1 395 174 2619
Email [email protected]
Background: MicroRNA-675-5p (miR-675-5p) is dysregulated in multiple human cancers, but its involvement in papillary thyroid cancer (PTC) remains to be investigated. This study aimed to examine the expression pattern of miR-675 in PTC, determine the effects of miR-675 on regulating the progression of PTC, and to explore the underlying molecular mechanisms.
Methods: The expression profile of miR-675 in PTC tissues and cell lines was determined using RT-qPCR. CCK-8, transwell migration and invasion assays, and xenograft tumors in nude mice were employed to analyze proliferation, in vitro migration and invasion, and in vivo tumor growth of PTC cells, respectively. The putative target of miR-675 was predicted using bioinformatic algorithms and was confirmed using luciferase reporter assays, RT-qPCR, and Western blotting.
Results: miR-675 expression was decreased in PTC tissues and cell lines. A low level of miR-675 expression was significantly correlated with lymphatic metastasis and TNM stage in PTC patients. Ectopic miR-675 expression suppressed PTC cell proliferation, migration, and invasion in vitro and hindered tumor growth in vivo. Mitogen-activated protein kinase 1 (MAPK1) was found to be the direct target gene of miR-675 in PTC cells. MAPK1 reintroduction negated the tumor-suppressing effect of miR-675 overexpression in PTC cells. Furthermore, the lncRNA mitochondrial RNA processing endoribonuclease (RMRP) functioned as a ceRNA of miR-675 in PTC cells. Silencing RMRP expression inhibited the growth and metastasis of PTC cells by sponging miR-675 and regulating MAPK1.
Conclusion: These findings revealed that miR-675 directly targets MAPK1 and is sponged by lncRNA RMRP to inhibit the oncogenicity of PTC, suggesting the RMRP-miR-675-MAPK1 pathway is an effective target for the treatment of PTC patients.
Keywords: papillary thyroid cancer, microRNA-675, mitogen-activated protein kinase 1, component of mitochondrial RNA processing endoribonuclease
Introduction
Thyroid cancer, the most common malignant endocrine tumor, accounts for about 2% of all newly diagnosed cases of cancers globally.1 The morbidity of thyroid cancer has been increasing year by year worldwide.2 Thyroid cancer can be divided into four major histological subtypes, including papillary thyroid cancer (PTC), follicular thyroid cancer, poorly differentiated carcinoma, and anaplastic thyroid cancer.3 PTC is the most prevalent histological subtype of thyroid cancer and accounts for approximately 85–90% of all thyroid cancer cases.4 Currently, thyroidectomy, in combination with radioiodine ablation and thyroid-stimulating hormone-suppressive therapy, is the primary treatment for patients with PTC.5 Most patients exhibit improved therapeutic outcomes after standard therapy; however, patients diagnosed at an advanced stage have a poorer prognosis.6 Therefore, elucidating the underlying molecular mechanisms that contribute to PTC pathogenesis and development is imperative for the identification of novel therapeutic techniques for the treatment of this disease.
MicroRNAs (miRNAs) are a series of non-coding short RNA molecules approximately 17–21 nucleotides long. They are implicated in the regulation of gene expression by directly interacting with partially complementary sequences in the 3′-untranslated regions (3ʹ-UTRs) of their target genes, which causes translational suppression and/or mRNA degradation.7 It is estimated that over one half of all miRNAs are located at cancer-related chromosomal regions, suggesting that miRNAs may play important roles in carcinogenesis and cancer progression.8–10 Numerous studies have emphasized the crucial roles of dysregulated miRNAs in the malignant progression of PTC.11–13 miRNAs are involved in the formation and progression of PTC by affecting numerous biological processes, such as cell proliferation, the cell cycle, apoptosis, and metastasis.11 miRNAs that are upregulated in PTC play oncogenic roles through the regulation of tumor suppressor genes,14,15 whereas miRNAs that are downregulated in PTC have tumor suppressor activity by directly targeting oncogenes.16,17 Accordingly, miRNAs may be attractive biomarkers for the diagnosing, treating, and predicting the prognosis of patients with PTC.
Long non-coding RNAs (lncRNAs) are members of the non-coding RNA family that are longer than 200 nucleotides and have no protein-coding function.18 An increasing number of studies have demonstrated that lncRNAs have important regulatory roles in nearly all cellular physiological and pathological processes.19–21 Specifically, lncRNAs are aberrantly expressed in PTC and their aberrant expression contributes to the aggressive behavior of PTC through their interactions with proteins, miRNAs, or mRNAs.22 Thus, exploring the influence of lncRNAs on the development of malignant PTC is essential for the development of effective treatment strategies for PTC.
miR-675-5p (miR-675) is dysregulated in multiple types of human cancer.23–27 However, the expression level, biological roles, and underlying mechanisms of miR-675 in PTC remain largely to be investigated. Therefore, in this study, the expression pattern of miR-675 in PTC was examined and the regulatory effects of miR-675 on PTC progression were determined by a series of in vitro experiments. Furthermore, the underlying molecular mechanisms of miR-675 in regulating PTC progression were also explored.
Materials and methods
Human tissue specimens
This study was approved by the Ethics Committee of JinLing Hospital and was performed in accordance with the Declaration of Helsinki. All participants provided written informed consent before surgical resection. PTC and adjacent normal tissues were collected from 57 patients who received surgery at JinLing Hospital. None of the patients enrolled in our current study had been previously treated with oncological surgery, chemotherapy, or radiotherapy. All tissues were snap-frozen in liquid nitrogen and transferred to −80 °C until further use.
Cell culture
Three PTC cell lines (HTH83, BCPAP, and TPC-1) and a normal human thyroid cell line (HT-ori3) were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% v/v heat-inactivated fetal bovine serum (FBS), 100 U/mL penicillin, and 100 mg/mL streptomycin (all from Invitrogen, Carlsbad, CA, USA). Cells were maintained at 37 °C in a humidified incubator supplemented with 5% CO2.
Transfection assays
miR-675 mimics, miRNA mimic negative control (miR-NC), miR-675 inhibitor and NC inhibitor were purchased from RiboBio (RiboBio, Guangzhou, China). Full-length MAPK1 sequences lacking the 3ʹ-UTR were chemically synthesized by GenePharma (Shanghai, China) and cloned into pcDNA3.1 plasmid to construct MAPK1 overexpression plasmid (pc-MAPK1). Small interfering RNAs (siRNAs) against MAPK1 (si-MAPK1) and RMRP (si-RMRP) and negative control siRNA (si-NC) were purchased from the Chinese Academy of Sciences (Changchun, China). Cells were plated onto 6-well plates at a density of 5×105 cells/well. After an overnight incubation, cells were transfected with miR-675 mimics (100 pmol), miR-NC (100 pmol), miR-675 inhibitor (100 pmol), NC inhibitor (100 pmol), pcDNA3.1 (4 μg), pc-MAPK1 (4 μg), si-MAPK1(100 pmol) or si-NC (100 pmol) using Lipofectamine 2000 reagents (Invitrogen), in accordance with the manufacturer’s protocol. Cells were then incubated at 37 °C under 5% CO2. After incubation 48 h, the transfection efficiency was evaluated using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Transfected cells were used in subsequent experiments after various incubation times.
Total RNA extraction and RT-qPCR
Total RNA was isolated from tissue specimens or cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. To quantify miR-675, first strand complementary DNA (cDNA) was synthesized from total RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The temperature protocol for reverse transcription was as follows: 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min. Subsequently, quantitative PCR (qPCR) was performed using a TaqMan MicroRNA PCR Kit (Applied Biosystems, Foster City, CA, USA), with U6 small nuclear as an internal reference. The temperature protocols for qPCR was as follows: 50 °C for 2 min, 95 °C for 10 min; 40 cycles of denaturation at 95 °C for 15 sec; and annealing/extension at 60 °C for 60 sec. To analyze MAPK1 mRNA and RMRP expression, total RNA was reverse transcribed into cDNA using a PrimeScript® RT reagent Kit, followed by qPCR using SYBR® Premix Ex TaqTM II (both from Takara Biotechnology CO., LTD., Dalian, China). The temperature protocol for reverse transcription was as follows: 37 °C for 15 min and 85 °C for 5 second. The qPCR was performed with cycling conditions as follows: 5 min at 95 °C, followed by 40 cycles of 95 °C for 30 sec and 65 °C for 45 sec. GAPDH was used as an endogenous control for normalizing MAPK1 and RMRP expression. Relative gene expression was calculated using the 2−ΔΔCt method.28
The primers were designed as follows: miR-675, 5′-UGGUGCGGAGAGGGCCCACAGUG-3′ (forward) and 5′-TGGTGTCGTGGAGTCG-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); RMRP, 5′-ACTCCAAAGTCCGCCAAGA-3′ (forward) and 5′-TGCGTAACTAGAGGGAGCTGAC-3′ (reverse); MAPK1, 5′-TGGATTCCCTGGTTCTCTCTAAAG-3′ (forward) and 5′-GGGTCTGTTTTCCGAGGATGA-3′ (reverse); and GAPDH, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse).
Cell counting kit-8 assay
Transfected cells were collected 24 h after incubation and were inoculated into 96-well plates at a density of 2,000 cells/well. A cell counting kit-8 (CCK-8) assay was used to detect cell proliferation at four time points subsequent to inoculation (0, 24, 48, and 72 h). Briefly, transfected cells were incubated with 10 μL of CCK-8 regent (Dojindo Laboratories, Kumamoto, Japan) for an additional 2 h. Absorbance was measured at 450 nm using a Bio-Rad iMark plate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell migration and invasion assays
Following 48 h of incubation, transfected cells were harvested, washed with PBS, and re-suspended in FBS-free DMEM. For invasion assays, 200 µL of FBS-free DMEM containing 1×105 transfected cells was seeded into the upper compartment of transwell chambers (Corning Incorporated, Corning, NY, USA) that were precoated with Matrigel (BD Biosciences, San Jose, CA, USA). The lower compartments were filled with 500 µL of DMEM containing 20% FBS. After 24 h of incubation, non-invading cells were gently removed using a cotton swab, while the invading cells were fixed in 100% methanol and stained with 0.5% crystal violet. Transwell migration assays were performed using an experimental procedure similar to the invasion assay, except that Matrigel was not used to coat the transwell chamber. Migratory and invasive capacities were determined by counting the number of cells that migrated or invaded in five representative microscopic fields under a light microscope (IX53; Olympus, Tokyo, Japan).
Xenograft tumors in nude mice
Nude BALB/c mice (female, 20 g, 4–5 weeks of age) were purchased from Vital River Laboratory Animal Technology (Beijing, China) and maintained in special pathogen-free conditions (25 °C; 50% humidity; 10-h light/14-h dark cycle). TPC-1 cells transfected with miR-675 mimics or miR-NC were collected after 24 h of incubation and then subcutaneously injected into nude mice (n=4 for each group). Two weeks later, the width and length of tumor xenografts that formed in nude mice were measured every 2 days using a Vernier caliper. All nude mice were sacrificed 4 weeks after cell implantation and tumor xenografts were resected and weighed. Tumor volume was calculated according to the formula: volume = (length × width2)/2. Animal experimental protocols were approved by the Animal Care Committee of the JinLing Hospital and were performed in accordance with the Animal Protection Law of the People’s Republic of China-2009.
Bioinformatic algorithms
LncBase Experimental version 2.0 (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2findex-experimental) was employed to predict the miR-675-RMRP axis. The putative targets of miR-675 were determined using TargetScan (www.targetscan.org) and miRanda (www.microrna.org).
Luciferase reporter assays
3′-UTR fragments of MAPK1 containing the predicted wild-type (wt) or mutant (mut) miR-675 binding site were amplified by GenePharma (Shanghai, China) and inserted into pMIR-REPOR (Promega Corp., Madison, WI, USA) to generate MAPK1-wt and MAPK1-mut plasmids, respectively. The luciferase reporter plasmids, RMRP-wt and RMRP-mut, were created in a similar manner. Cells were maintained in 24-well plates and co-transfected with the constructed luciferase reporter plasmids and miR-675 mimics or miR-NC using Lipofectamine 2000, as per the manufacturer’s instructions. The luciferase activity in the cell lysates was detected 48 h after transfection, using a dual-luciferase reporter assay system (Promega Corp.). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western blotting analysis
Total protein was extracted from cultured cells using RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA) and then quantified using a BCATM Protein Assay Kit (Thermo Fisher Scientific). Equal amounts of protein were separated on 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes (Beyotime Institute of Biotechnology, Shanghai, China). Following blocking by incubation with 5% skimmed milk, membranes were incubated overnight at 4 °C with mouse antibodies against MAPK1 (1:1,000 dilution; sc-81459; Santa Cruz Biotechnology Inc., Dallas, TX, USA) or GAPDH (1:1,000 dilution; sc-32233; Santa Cruz Biotechnology Inc.), followed by further incubation with goat anti-mouse horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; sc-516132; Santa Cruz Biotechnology Inc.) for 2 h at room temperature. Protein signals were visualized using an Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA, USA). GAPDH served as an internal control.
Statistical analysis
Data are shown as mean ± standard deviation. All statistical analyses were performed using a Student’s t-test when only two groups were compared and using a one-way analysis of variance (ANOVA) when three or more groups were compared. A Student-Newman-Keuls test was used as a post hoc test after ANOVA. The association between miR-675 and clinicopathological parameters in patients with PTC was examined using a Chi square test. The relationship between miR-675 and MAPK1 mRNA levels was analyzed by Spearman’s correlation analysis. SPSS (version 17.0; IBM Corporation, USA) was used for all statistical analyses and P<0.05 was considered statistically significant for all analyses.
Results
The expression profile of miR-675 in PTC and its correlation with clinicopathological parameters
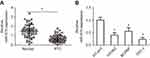
To determine the role of miR-675 in PTC, we first measured miR-675 expression in 57 pairs of PTC and adjacent normal tissues using RT-qPCR. miR-675 was clearly downregulated in PTC tissues compared to adjacent normal tissues (Figure 1A, P<0.05). In addition, we also examined the expression of miR-675 in different PTC cell lines. The expression level of miR-675 was lower in all three PTC cell lines (HTH83, BCPAP, and TPC-1) relative to its expression in a normal human thyroid cell line (HT-ori3; Figure 1B, P<0.05).
Based on the median miR-675 expression level in PTC tissues, all patients with PTC were divided into two groups: high miR-675 expression group (miR-675 expression above the median value) and low miR-675 expression group (miR-675 expression below the median value). The association between miR-675 expression and clinicopathological parameters in patients with PTC was explored and the results are shown in Table 1. Lower levels of miR-675 expression were correlated with lymphatic metastasis (P=0.001) and TNM stage (P=0.007) in patients with PTC. These observations implied that the aberrant downregulation of miR-675 may affect the malignancy of PTC.
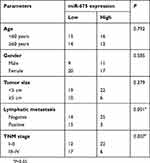 |
Table 1 The association between miR-675 and clinicopathological parameters in patients with PTC |
miR-675 suppressed the growth and metastasis of HTH83 and TPC-1 cells in vitro
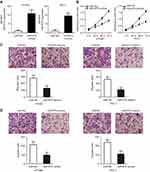
To explore the function of miR-675 in PTC progression, HTH83 and TPC-1 cells, which express relatively low levels of miR-675 among the three PTC cell lines used in this study, were transfected with miR-675 mimics or miR-NC. RT-qPCR analysis demonstrated that miR-675 was markedly overexpressed in HTH83 and TPC-1 cells after transfection with miR-675 mimics (Figure 2A, P<0.05). CCK-8 assays were performed to evaluate the effects of miR-675 on the proliferation of PTC cells. Ectopic miR-675 expression resulted in a clear decrease in the proliferation of HTH83 and TPC-1 cells when compared with the proliferation of cells transfected with miR-NC (Figure 2B, P<0.05). Transwell migration and invasion assays indicated that miR-675 expression inhibited the migratory (Figure 2C, P<0.05) and invasive (Figure 2D, P<0.05) abilities of HTH83 and TPC-1 cells. These results demonstrated that miR-675 exhibited inhibitory effects on the growth and metastasis of PTC cells.
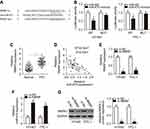
miR-675 directly targeted MAPK1 in PTC cells
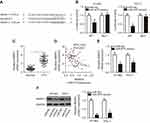
To clarify the mechanisms underlying the activity of miR-675 in PTC cells, bioinformatic algorithms (TargetScan and miRanda) were utilized to predict the potential targets of miR-675. Among these candidates, MAPK1 was chosen for further investigation because it is implicated in the pathogenesis of PTC (Figure 3A).29,30 Luciferase reporter assays were then performed to determine whether the 3′-UTR of MAPK1 could be directly targeted by miR-675 in PTC cells. HTH83 and TPC-1 cells were transiently co-transfected with MAPK1-wt or MAPK1-mut, and miR-675 mimics or miR-NC. After transfection, luciferase reporter assays were performed. Upregulation of miR-675 resulted in the significant downregulation of MAPK1-wt luciferase activity in HTH83 and TPC-1 cells (Figure 3B, P<0.05), but did not affect MAPK1-mut luciferase activity, suggesting that miR-675 could recognize and bind to the 3′-UTR of MAPK1 in PTC cells.
To further examine the correlation between miR-675 and MAPK1 in PTC, RT-qPCR analysis was performed to measure MAPK1 mRNA expression in PTC tissues. MAPK1 expression was found to be significantly upregulated in PTC tissues (Figure 3C, P<0.05). In addition, Spearman’s correlation analysis demonstrated an inverse correlation between miR-675 and MAPK1 mRNA expression in PTC tissues (Figure 3D; R2=0.2823, P<0.0001). Furthermore, RT-qPCR and Western blotting analysis found that miR-675 decreased MAPK1 expression in HTH83 and TPC-1 cells at both the mRNA (Figure 3E, P<0.05) and protein (Figure 3F, P<0.05) levels. These results demonstrated that MAPK1 is a direct target gene of miR-675 in PTC cells.
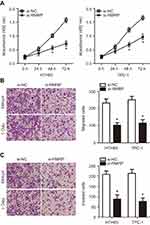
MAPK1 reintroduction impaired miR-675 mimics-induced suppression of PTC cell proliferation, migration and invasion
To examine the functional relevance of MAPK1 targeting by miR-675, we explored whether restoration of MAPK1 could abrogate the tumor suppressor activity of miR-675 in PTC cells. HTH83 and TPC-1 cells with high levels of miR-675 expression were transfected with the MAPK1 overexpression plasmid, pc-MAPK1, or an empty pcDNA3.1 plasmid. Western blotting analysis showed that MAPK1 protein expression that was reduced by the upregulation of miR-675, was recovered by pc-MAPK1 co-transfection (Figure 4A, P<0.05). Next, functional experiments indicated that miR-675 overexpression significantly suppressed the proliferation (Figure 4B, P<0.05), migration (Figure 4C, P<0.05), and invasion (Figure 4D, P<0.05) of HTH83 and TPC-1 cells, whereas these suppressive effects were rescued by the restoration of MAPK1 expression. These results clearly showed that miR-675 inhibited the malignant phenotypes of PTC cells, at least partly, by decreasing MAPK1 expression.
LncRNA RMRP functioned as a sponge of miR-675 in PTC cells
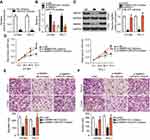
LncRNAs are known as to act as competing endogenous RNAs (ceRNAs) for miRNAs.31 To determine whether miR-675 could be sponged by certain lncRNAs, bioinformatic analysis was performed, which identified a potential miR-675 binding site in RMRP (Figure 5A). RMRP was selected for further analysis, as RMRP has been found to be closely related with the carcinogenesis and cancer progression.32–35 Luciferase reporter assays were then performed to determine whether miR-675 targeted RMRP in PTC. The RMRP-wt and RMRP-mut reporter plasmids, along with miR-675 mimics or miR-NC, were transfected into HTH83 and TPC-1 cells. Transfection with miR-675 mimics dramatically suppressed the luciferase activity of RMRP-wt in HTH83 and TPC-1 cells (Figure 5B, P<0.05), but the luciferase activity of RMRP-mut was not affected by miR-675 upregulation.
To confirm these findings, we first measured RMRP expression in PTC and assessed its relationship with miR-675. The expression of RMRP was found to be elevated in PTC tissues, compared with adjacent normal tissues (Figure 5C, P<0.05). Furthermore, RMRP expression was inversely correlated with miR-675 expression in PTC tissues (Figure 5D; R2=0.3447, P<0.0001). In addition, treatment with siRNA against RMRP (si-RMRP) decreased RMRP expression (Figure 5E, P<0.05) and subsequently, increased miR-675 expression (Figure 5F, P<0.05) in both HTH83 and TPC-1 cells, as indicated by RT-qPCR analysis. Furthermore, Western blotting analysis showed that silencing RMRP expression significantly decreased MAPK1 protein expression in HTH83 and TPC-1 cells (Figure 5G, P<0.05). Collectively, our data suggested that RMRP directly interacted with miR-675 and regulated MAPK1 expression, possibly by acting as a ceRNA.
Downregulation of lncRNA RMRP inhibited the proliferation, migration, and invasion of HTH83 and TPC-1 cells
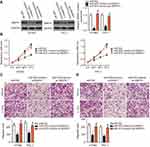
To investigate the detailed role of RMRP in the malignancy of PTC, si-RMRP or si-NC were transfected into HTH83 and TPC-1 cells and a series of functional experiments were then performed. CCK-8 assays indicated that the downregulation of RMRP effectively inhibited the proliferation of HTH83 and TPC-1 cells (Figure 6A, P<0.05). Moreover, RMRP knockdown restricted the migration (Figure 6B, P<0.05) and invasion (Figure 6C, P<0.05) of HTH83 and TPC-1 cells. These results demonstrated that RMRP knockdown led to the suppression of cell growth and metastasis in PTC.
Decreasing RMRP expression inhibited the growth and metastasis of HTH83 and TPC-1 cells by sponging mir-675and regulating MAPK1 expression
To determine whether miR-675 can functionally rescue RMRP function in PTC cells, RMRP decreasing-HTH83 and TPC-1 cells were co-transfected with miR-675 inhibitor or NC inhibitor. Firstly, RT-qPCR analysis showed that transfection of miR-675 inhibitor efficiently silenced miR-675 expression in HTH83 and TPC-1 cells (Figure 7A, P<0.05). Moreover, the upregulation of miR-675 (Figure 7B, P<0.05) and the downregulation of MAPK1 protein (Figure 7C, P<0.05) in HTH83 and TPC-1 cells caused by RMRP knockdown was recovered after co-transfection of miR-675 inhibitor. Furthermore, functional experiments indicated that inhibition of miR-675 partially neutralized the influence of RMRP knockdown on the proliferation (Figure 7D, P<0.05), migration (Figure 7E, P<0.05), and invasion (Figure 7F, P<0.05) of HTH83 and TPC-1 cells. Taken together, these results suggested that inhibition of RMRP could prevent PTC progression by targeting the miR-675/MAPK1 axis.
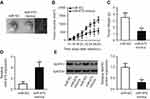
miR-675 inhibited tumor growth in vivo
We next studied the effect of miR-675 upregulation on xenograft tumor growth of PTC cells in nude mice. Tumor volume was reduced in nude mice inoculated with miR-675 compared with those inoculated with TPC-1 cells expressing miR-NC (Figure 8A and B, P<0.05). Meanwhile, tumor weight in the miR-675 mimic group was significantly less than in the miR-NC group (Figure 8C, P<0.05). In addition, miR-675 expression was detected in dissected tumor xenografts and miR-675 expression remained upregulated in tumor xenografts derived from miR-675 mimic-transfected TPC-1 cells (Figure 8D, P<0.05). Furthermore, the protein expression of MAPK1 in tumor xenografts was examined using Western blotting. MAPK1 protein levels were lower in miR-675-overexpressing tumor xenografts (Figure 8E, P<0.05). In summary, these findings revealed that miR-675 was able to hinder PTC tumor growth in vivo.
Discussion
Recent studies have demonstrated that numerous miRNAs are dysregulated in PTC.36–39 The dysregulated miRNAs and their direct target genes consist of a complex network, which are implicated in the initiation and progression of PTC by regulating a range of biological processes.40–42 Hence, further investigation on the biological roles of dysregulated miRNAs in PTC may contribute toward the development of effective therapeutic targets for patients with PTC. To the best of our knowledge, this is the first study investigating the role of miR-675 in PTC.
miR-675 is upregulated in multiple types of human cancer. For instance, miR-675 is highly expressed in breast cancer and its level of expression is significantly correlated with tumor grade.23 miR-675 is also overexpressed in head and neck squamous cell carcinoma,24 bladder cancer,25 and hepatocellular carcinoma.26,27 In contrast, miR-675 is expressed at low levels in non-small cell lung cancer,43 adrenocortical adenoma,44 prostate cancer,45 pancreatic cancer,46 and glioma.47 However, the expression status of miR-675 in PTC remains unknown. In this study, we used RT-qPCR to investigate the expression pattern of miR-675 in PTC. miR-675 was found to be downregulated in both PTC tissues and cell lines. Decreased miR-675 expression presented an obvious association with lymphatic metastasis and TNM stage in PTC patients. These findings suggest that miR-675 is a potential biomarker for the diagnosis and prognosis of PTC. However, in this study, we did not use TCGA database to determine the expression profile of miR-675 in PTC and identify its association with clinical parameters of patients with PTC. It was a limitation of our study, and we will resolve it in the near future.
miR-675 overexpression increases cell proliferation and migration in vitro and promotes tumor growth and metastasis in vivo.48 In bladder cancer, miR-675 inhibition attenuates cell proliferation and induces cell cycle arrest and apoptosis.25 miR-675 also acts as an oncogene in colon cancer49 and hepatocellular carcinoma.26,27 In contrast, miR-675 has been identified as a tumor suppressor in non-small cell lung cancer by affecting cell proliferation, colony formation, and metastasis in vitro and tumor growth in vivo.43 In pancreatic cancer, miR-675 upregulation suppresses cell growth and metastasis and increase apoptosis in vitro.46 miR-675 also plays tumor-suppressing roles in prostate cancer45 and glioma.47 However, the detailed roles of miR-675 in the development of PTC remain unclear. In the current study, functional analysis showed that enforced miR-675 expression repressed PTC cell proliferation, migration and invasion in vitro as well as hindered tumor growth in vivo. These findings suggest that miR-675 is a potential therapeutic target in patients with PTC.
A variety of genes, including p53,25 Cdc25A,26 AKT,27 GPR55,43 TGFBI,45 ZEB1,46 CDK6,47 c-Cbl,48 and Cbl-b,48 have been shown to be direct targets of miR-675. MAPK1, a member of the mitogen activated protein kinase signaling cascade, was identified as a novel direct target of miR-675 in PTC cells. RMRP acted as a ceRNA to modulate MAPK1 expression by sponging miR-675. MAPK1 is a well-known oncogene and is overexpressed in various types of human cancer, such as lung cancer,50 ovarian cancer,51,52 cervical cancer,53 gastric cancer,54 myeloma,55 and sacral chordoma.56 Expression of MAPK1 is also increased in PTC and the deregulation of MAPK1 plays a crucial role in the development of PTC by regulating important pathological processes.29,30 Here, we showed that miR-675 directly targets MAPK1 to inhibit PTC progression in vitro and in vivo. Accordingly, miR-675-mediated inhibition of MAPK1 may be an effective therapeutic technique for PTC patients in the future.
RMRP is upregulated in bladder cancer and its downregulation is inversely associated with tumor size and lymph node metastasis.32 Bladder cancer patients with high levels of RMRP expression have a shorter survival period than patients with low levels of RMRP expression.32 RMRP expression is increased in gastric cancer and increased RMRP expression is correlated with Borrmann classification and metastasis.33 RMRP is also highly expressed in neonatal neuroblastoma34 and lung cancer.35 RMRP exerts oncogenic roles in cancer pathogenesis and progression via different mechanisms depending on the cancer type.32–35 Here, we demonstrated that RMRP inhibition suppressed the oncogenicity of PTC by sponging miR-675 and regulating MAPK1 expression. Hence, targeting RMRP, which may result in miR-675 upregulation and MAPK1 downregulation, may be an attractive therapeutic technique for patients with PTC.
Conclusion
In summary, our studies showed that miR-675 is downregulated in PTC tissues and cell lines. Downregulation of miR-675 was closely associated with poor prognosis in patients with PTC. miR-675 directly targets MAPK1 and is sponged by lncRNA RMRP to inhibit the malignancy of PTC in vitro and in vivo. These findings may provide a novel mechanism for PTC pathogenesis and suggest that miR-675 is a promising therapeutic target for patients with this disease.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nikiforova MN, Nikiforov YE. Molecular genetics of thyroid cancer: implications for diagnosis, treatment and prognosis. Expert Rev Mol Diagn. 2008;8(1):83–95. doi:10.1586/14737159.8.1.83
2. Du L, Wang Y, Sun X, et al. Thyroid cancer: trends in incidence, mortality and clinical-pathological patterns in Zhejiang Province, Southeast China. BMC Cancer. 2018;18(1):291. doi:10.1186/s12885-018-4242-8
3. Xing M, Haugen BR, Schlumberger M. Progress in molecular-based management of differentiated thyroid cancer. Lancet. 2013;381(9871):1058–1069. doi:10.1016/S0140-6736(13)60109-9
4. Peng XG, Chen ZF, Zhang KJ, et al. VEGF Trapon inhibits tumor growth in papillary thyroid carcinoma. Eur Rev Med Pharmacol Sci. 2015;19(2):235–240.
5. Nikiforov YE, Nikiforova MN. Molecular genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol. 2011;7(10):569–580.
6. Voutilainen PE, Multanen MM, Leppaniemi AK, Haglund CH, Haapiainen RK, Franssila KO. Prognosis after lymph node recurrence in papillary thyroid carcinoma depends on age. Thyroid. 2001;11(10):953–957.
7. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531.
8. Gulino R, Forte S, Parenti R, Memeo L, Gulisano M. MicroRNA and pediatric tumors: future perspectives. Acta Histochem. 2015;117(4–5):339–354.
9. Ma XP, Zhang T, Peng B, Yu L, Jiang de K. Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS One. 2013;8(11):e79584.
10. Calabrese G, Dolcimascolo A, Torrisi F, Zappala A, Gulino R, Parenti R. MiR-19a overexpression in FTC-133 cell line induces a more de-differentiated and aggressive phenotype. Int J Mol Sci. 2018;19(12):3944. doi:10.3390/ijms19123944
11. Aragon Han P, Weng CH, Khawaja HT, et al. MicroRNA expression and association with clinicopathologic features in papillary thyroid cancer: a systematic review. Thyroid. 2015;25(12):1322–1329. doi:10.1089/thy.2015.0193
12. Hu Y, Wang H, Chen E, Xu Z, Chen B, Lu G. Candidate microRNAs as biomarkers of thyroid carcinoma: a systematic review, meta-analysis, and experimental validation. Cancer Med. 2016;5(9):2602–2614. doi:10.1002/cam4.811
13. Zhu G, Xie L, Miller D. Expression of MicroRNAs in thyroid carcinoma. Methods Mol Biol. 2017;1617:261–280. doi:10.1007/978-1-4939-7046-9_19
14. Huang Y, Yu S, Cao S, et al. MicroRNA-222 promotes invasion and metastasis of papillary thyroid cancer through targeting protein phosphatase 2 regulatory subunit B alpha expression. Thyroid. 2018. doi:10.1089/thy.2017.0665
15. Fang L, Kong D, Xu W. MicroRNA-625-3p promotes the proliferation, migration and invasion of thyroid cancer cells by up-regulating astrocyte elevated gene 1. Biomed Pharmacother. 2018;102:203–211. doi:10.1016/j.biopha.2018.03.043
16. Fu YT, Zheng HB, Zhang DQ, Zhou L, Sun H. MicroRNA-1266 suppresses papillary thyroid carcinoma cell metastasis and growth via targeting FGFR2. Eur Rev Med Pharmacol Sci. 2018;22(11):3430–3438. doi:10.26355/eurrev_201806_15166
17. Li R, Dong B, Wang Z, Jiang T, Chen G. MicroRNA-361-5p inhibits papillary thyroid carcinoma progression by targeting ROCK1. Biomed Pharmacother. 2018;102:988–995. doi:10.1016/j.biopha.2018.03.122
18. Lanzafame M, Bianco G, Terracciano LM, Ng CKY, Piscuoglio S. The role of long non-coding RNAs in hepatocarcinogenesis. Int J Mol Sci. 2018;19(3):682. doi:10.3390/ijms19030682
19. He RZ, Luo DX, Mo YY. Emerging roles of lncRNAs in the post-transcriptional regulation in cancer. Genes Dis. 2019;6(1):6–15. doi:10.1016/j.gendis.2019.01.003
20. Youness RA, Gad MZ. Long non-coding RNAs: functional regulatory players in breast cancer. Non-coding RNA Res. 2019;4(1):36–44. doi:10.1016/j.ncrna.2019.01.003
21. Li Y, Yang X, Kang X, Liu S. The regulatory roles of long noncoding RNAs in the biological behavior of pancreatic cancer. Saudi J Gastroenterol. 2019;25(3):145–151.
22. Murugan AK, Munirajan AK, Alzahrani AS. Long noncoding RNAs: emerging players in thyroid cancer pathogenesis. Endocr Relat Cancer. 2018;25(2):R59–R82. doi:10.1530/ERC-17-0188
23. Zhai LL, Wang P, Zhou LY, et al. Over-expression of miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues of breast cancer patients. Int J Clin Exp Med. 2015;8(7):11195–11201.
24. Guan GF, Zhang DJ, Wen LJ, et al. Overexpression of lncRNA H19/miR-675 promotes tumorigenesis in head and neck squamous cell carcinoma. Int J Med Sci. 2016;13(12):914–922. doi:10.7150/ijms.16571
25. Liu C, Chen Z, Fang J, Xu A, Zhang W, Wang Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016;37(1):263–270. doi:10.1007/s13277-015-3779-2
26. Yu YQ, Weng J, Li SQ, Li B, Lv J. MiR-675 promotes the growth of hepatocellular carcinoma cells through the Cdc25A pathway. Asian Pac J Cancer Prev. 2016;17(8):3881–3885.
27. Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3beta/Cdc25A signaling pathway. J Huazhong Univ Sci Technol Med Sci. 2014;34(3):363–369. doi:10.1007/s11596-014-1284-2
28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
29. Wang J, Yang H, Si Y, et al. Iodine promotes tumorigenesis of thyroid cancer by suppressing Mir-422a and up-regulating MAPK1. Cell Physiol Biochem. 2017;43(4):1325–1336. doi:10.1159/000481844
30. Hanly EK, Tuli NY, Bednarczyk RB, et al. Hyperactive ERK and persistent mTOR signaling characterize vemurafenib resistance in papillary thyroid cancer cells. Oncotarget. 2016;7(8):8676–8687. doi:10.18632/oncotarget.6779
31. Fernandes JCR, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long non-coding RNAs in the regulation of gene expression: physiology and disease. Non-coding RNA. 2019;5(1):17. doi:10.3390/ncrna5010017
32. Cao HL, Liu ZJ, Huang PL, Yue YL, Xi JN. lncRNA-RMRP promotes proliferation, migration and invasion of bladder cancer via miR-206. Eur Rev Med Pharmacol Sci. 2019;23(3):1012–1021. doi:10.26355/eurrev_201902_16988
33. Shao Y, Ye M, Li Q, et al. LncRNA-RMRP promotes carcinogenesis by acting as a miR-206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7(25):37812–37824. doi:10.18632/oncotarget.9336
34. Pan J, Zhang D, Zhang J, Qin P, Wang J. LncRNA RMRP silence curbs neonatal neuroblastoma progression by regulating microRNA-206/tachykinin-1 receptor axis via inactivating extracellular signal-regulated kinases. Cancer Biol Ther. 2019;20(5):653–665.
35. Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One. 2016;11(12):e0164845. doi:10.1371/journal.pone.0164845
36. Chou CK, Liu RT, Kang HY. MicroRNA-146b: a novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci. 2017;18(3):636. doi:10.3390/ijms18030636
37. Zhang M, Wu W, Gao M, Fei Z. MicroRNA-451 as a prognostic marker for diagnosis and lymph node metastasis of papillary thyroid carcinoma. Cancer Biomarkers. 2017;19(4):437–445. doi:10.3233/CBM-170059
38. Xue KC, Hu DD, Zhao L, Li N, Shen HY. MiR-577 inhibits papillary thyroid carcinoma cell proliferation, migration and invasion by targeting SphK2. Eur Rev Med Pharmacol Sci. 2017;21(17):3794–3800.
39. Wang R, Ma Q, Ji L, Yao Y, Ma M, Wen Q. miR-622 suppresses tumor formation by directly targeting VEGFA in papillary thyroid carcinoma. Onco Targets Ther. 2018;11:1501–1509. doi:10.2147/OTT.S156810
40. Zhang X, Mao H, Lv Z. MicroRNA role in thyroid cancer pathogenesis. Front Biosci. 2013;18:734–739. doi:10.2741/4135
41. de la Chapelle A, Jazdzewski K. MicroRNAs in thyroid cancer. J Clin Endocrinol Metab. 2011;96(11):3326–3336. doi:10.1210/jc.2011-1004
42. Menon MP, Khan A. Micro-RNAs in thyroid neoplasms: molecular, diagnostic and therapeutic implications. J Clin Pathol. 2009;62(11):978–985. doi:10.1136/jcp.2008.063909
43. He D, Wang J, Zhang C, et al. Down-regulation of miR-675-5p contributes to tumor progression and development by targeting pro-tumorigenic GPR55 in non-small cell lung cancer. Mol Cancer. 2015;14:73. doi:10.1186/s12943-014-0278-9
44. Schmitz KJ, Helwig J, Bertram S, et al. Differential expression of microRNA-675, microRNA-139-3p and microRNA-335 in benign and malignant adrenocortical tumours. J Clin Pathol. 2011;64(6):529–535. doi:10.1136/jcp.2010.085621
45. Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. Febs J. 2014;281(16):3766–3775. doi:10.1111/febs.12902
46. Wang J, Zhang Y, Wei H, et al. The mir-675-5p regulates the progression and development of pancreatic cancer via the UBQLN1-ZEB1-mir200 axis. Oncotarget. 2017;8(15):24978–24987. doi:10.18632/oncotarget.15330
47. Li C, Lei B, Huang S, et al. H19 derived microRNA-675 regulates cell proliferation and migration through CDK6 in glioma. Am J Transl Res. 2015;7(10):1747–1764.
48. Vennin C, Spruyt N, Dahmani F, et al. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget. 2015;6(30):29209–29223.
49. Costa V, Lo Dico A, Rizzo A, et al. MiR-675-5p supports hypoxia induced epithelial to mesenchymal transition in colon cancer cells. Oncotarget. 2017;8(15):24292–24302.
50. You B, Yang YL, Xu Z, et al. Inhibition of ERK1/2 down-regulates the Hippo/YAP signaling pathway in human NSCLC cells. Oncotarget. 2015;6(6):4357–4368.
51. Yiwei T, Hua H, Hui G, Mao M, Xiang L. HOTAIR interacting with MAPK1 regulates ovarian cancer skov3 cell proliferation, migration, and invasion. Med Sci Monit. 2015;21:1856–1863.
52. Rahman MT, Nakayama K, Rahman M, et al. KRAS and MAPK1 gene amplification in type II ovarian carcinomas. Int J Mol Sci. 2013;14(7):13748–13762.
53. Li XW, Tuergan M, Abulizi G. Expression of MAPK1 in cervical cancer and effect of MAPK1 gene silencing on epithelial-mesenchymal transition, invasion and metastasis. Asian Pac J Trop Med. 2015;8(11):937–943.
54. Fei B, Wu H. MiR-378 inhibits progression of human gastric cancer MGC-803 cells by targeting MAPK1 in vitro. Oncol Res. 2012;20(12):557–564.
55. Tsubaki M, Takeda T, Ogawa N, et al. Overexpression of survivin via activation of ERK1/2, Akt, and NF-kappaB plays a central role in vincristine resistance in multiple myeloma cells. Leuk Res. 2015;39(4):445–452.
56. Zhang K, Chen H, Zhang B, et al. Overexpression of Raf-1 and ERK1/2 in sacral chordoma and association with tumor recurrence. Int J Clin Exp Pathol. 2015;8(1):608–614.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.