Back to Journals » OncoTargets and Therapy » Volume 11
MicroRNA-645 targets urokinase plasminogen activator and decreases the invasive growth of MDA-MB-231 triple-negative breast cancer cells
Authors Meng D, Lei M , Han Y, Zhao D, Zhang X , Yang Y , Liu R
Received 11 September 2018
Accepted for publication 8 October 2018
Published 2 November 2018 Volume 2018:11 Pages 7733—7743
DOI https://doi.org/10.2147/OTT.S187221
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Du Meng,1,* Ming Lei,2,* Yaxuan Han,3 Dongli Zhao,2 Xiaozhi Zhang,1 Yunyi Yang,1 Rui Liu1
1Department of Radio Oncology, The First Affiliated Hospital of Xi’an Jiaotong University School of Medicine, Xi’an 710061, Shaanxi Province, People’s Republic of China; 2Department of Cardiothoracic Surgery, The NO 3 Hospital of Xi’an, Xi’an 710000, Shaanxi Province, People’s Republic of China; 3Department of Oncology, The Xi’an Chest Hospital, Xi’an 710000, Shaanxi Province, People’s Republic of China
*These authors contributed equally to this work
Background: Urokinase plasminogen activator (uPA) promotes the in vivo invasive growth of HCC cells by cleaving and activating matrix metalloproteinases (MMPs) to induce the destruction of the extracellular matrix of triple-negative breast cancer (TNBC) cells. The identification of microRNAs that target uPA and decrease uPA expression would be useful for attenuating the in vivo invasive growth of TNBC cells.
Materials and methods: MicroRNA-645 (miR-645) was identified using an online tool (miRDB) as potentially targeting uPA; miR-645 inhibition of uPA was confirmed by western blot experiments. The effects of miR-645 on the in vivo invasive growth of TNBC cells were examined using an intrahepatic tumor model in nude mice, and the miR-645 mechanism of action was explored with MMP cleaving experiments.
Results: Through virtual screening, we discovered that miR-645 potentially targeted the uPA 3' untranslated region. This targeting was confirmed by western blot experiments and miR-645 lentiviral particle (LV-645) transduction that inhibited uPA expression in MDA-MB-231 TNBC cells. The LV-645 inhibition of uPA led to the decreased invasive growth of TNBC cells in nude mice. The mechanism data indicated that the uPA inhibition resulted in a decreased cleaving of the pro-MMP-9 protein.
Conclusion: Targeting uPA with miR-645 decreased the in vivo invasive growth of TNBC cells. These results suggest that miR-645 may represent a promising treatment strategy for TNBC.
Keywords: triple-negative breast cancer, urokinase plasminogen activator, microRNA, invasive growth
Introduction
Breast cancer represents one of the foremost threats to women’s health.1,2 There are three major subtypes of human breast cancer: endocrine-related/dependent breast cancer (estrogen receptor positive),3,4 human epidermal growth factor receptor 2 (HER2)-positive breast cancer5–7 and triple-negative breast cancer (TNBC) (estrogen receptor negative, progesterone receptor negative and HER2 negative).8,9 While there are effective treatment strategies for endocrine-related/dependent breast cancer (tamoxifen and fulvestrant),10–13 and antibody treatments (trastuzumab)14–16 can effectively attenuate the progression of HER2-positive breast cancer, there is no effective therapeutic intervention for TNBC. Therefore, it is important to identify novel therapeutic molecular targets for TNBC treatment.17,18
Urokinase plasminogen activator (uPA), which is encoded by the PLAU gene, is an important serine protease that mediates the conversion of inactive plasminogen to active plasmin.19–22 In normal cells, uPA expression is minimal, but in tumor cells, uPA expression increases severalfold.23 Recently, uPA has been recognized as one of the key regulators of the metastatic processes related to human cancer.24 With a high degree of substrate specificity, uPA cleaves and activates the matrix metalloproteinases (MMPs) that break down the extracellular matrix (ECM) of cancer cells and, in turn, promote the invasion and migration of human cancers.25,26 Inhibition of uPA represents a promising approach to the prevention and treatment of human cancer metastasis.
MicroRNAs (miRs) are a series of non-coding RNAs in mammalian cells that function as post-transcription regulators by targeting the 3′ untranslated region (UTR) of the targeted gene’s mRNA.27–30 Recent research has focused on miRs as a promising approach to inhibit the proliferation, invasion and migration of human cancer cells.31–34 In the present work, an online tool (miRDB) identified miR-645 as potentially targeting uPA; miR-645 received the software program’s highest score. Infection of miR-645 lentiviral particle (LV-645) significantly decreased uPA expression. Transfection of an uPA expression vector with either a mutated miR-645 target sequence or a miR-645 inhibitor almost blocked the inhibitory effect of LV-645 on uPA expression. LV-645 infection significantly decreased uPA expression and inhibited the cleaving of pro-MMP-9. The in vivo data showed that LV-645 significantly inhibited the invasive growth of MDA-MB-231 cells in the liver organs of nude mice.
Material and methods
Plasmids and reagents
The expression vector for uPA (PLAU gene) (Cat. No CH820007) was purchased from Vigene Corporation, Jinan City, Shandong Province, People’s Republic of China. The expression vector for the uPA lentivirus particles with the mutated miR-645 target sequences in the 3′ UTR was constructed by Vigene Corporation; the miR-645 lentivirus particles were also purchased from Vigene Corporation. The miR-645 inhibitor (Cat. No AM17100-MH11622) was purchased from Thermo Fisher Corporation, Waltham, MA, USA. The breast cancer cell lines, ZR75-1 cells (an endocrine-related breast cancer/estrogen receptor-positive breast cancer cell line), BT474 and SK-BR3 cells (two HER2-positive breast cancer cell lines) were purchased from Type Culture Collection of the Chinese Academy of Sciences, Shanghai, People’s Republic of China, a culture collection center of the Chinese government. The MDA-MB-231 cells (a metastatic TNBC cell line) and MCF-7 (an endocrine-related breast cancer/estrogen receptor-positive breast cancer cell line) were purchased from the Type Culture Collection of the Chinese Academy of Sciences and were gifts from Dr Fan Feng of the Research Center for Clinical and Translational Medicine, the 302nd Hospital of Chinese PLA, Beijing 100039, People’s Republic of China.35,36 The cells were preserved in our lab and maintained in DMEM (Thermo Fisher Corporation) with 10% FBS (Thermo Fisher Corporation) at 37°C with 5% CO2. All experiments, protocols and usage of cell lines were approved by Ethics Committee of the First Affiliated Hospital of Xi’an Jiao Tong University.
Antibodies and western blot
Antibodies to uPA (Cat. No ab169754), CYP3A4 (Cat. No ab155029), P-glycoprotein (Cat. No ab235954) and β-actin (Cat. No ab205) were obtained from Abcam Corporation, Cambridge, MA, USA. Protein A magnetic beads (Cat. No ab214286) linked with a secondary antibody (goat to rabbit) were also purchased from Abcam Corporation. The MDA-MB-231 cells that had been infected with control miRNA, LV-645, LV-645+ uPAMut or LV-645+ miR-645’s inhibitor were harvested to extract total protein samples for the western blot experiments. The western blot experiments were performed following the standard western blot protocol. The uPA expression in the MDA-MB-231 cells was measured using the uPA-specific antibodies.
Immunoprecipitation and biochemical cleavage assay
The MDA-MB-231 cells that were infected with either LV-645 or the control miRNA were harvested and lysed in the immunoprecipitation buffer 18–24 hours after culture at 4°C. The co-immunoprecipitation analysis was performed following methods that have been previously described in the literature.37,38 Briefly, the cell lysate was incubated with the anti-uPA antibodies and the Protein A magnetic beads that had been linked with a secondary antibody (goat to rabbit). The uPA proteins in the MDA-MB-231 cell lysate were isolated by the Protein A magnetic beads. The amount of uPA in the cell lysate (input) or attached to the Protein A magnetic beads (immunoprecipitated uPA) was examined using western blot experiments.
The biochemical cleavage assay that was used to measure the uPA cleavage of pro-MMP-9 was performed as described by Zhao et al, 2008.39 The recombinant pro-MMP-9 protein was purified and isolated by our lab using the methods that have been previously described in the literature.39,40 Protein A magnetic beads with attached uPA proteins were incubated with pro-MMP-9 protein (0.005 mol/L).39,40 Silver staining was used to examine the amounts of pro-MMP-9 and activated MMP-9 (act-MMP-9).
In vivo tumor growth (subcutaneous TNBC tumor model)
The animal experiments were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Xi’an Jiao Tong University. All the animal studies were conducted in accordance with the U.K. Animals (Scientific Procedures) Act of 1986 and the associated guidelines. The nude mice were purchased from Si-Bei-Fu Biotechnology Corporation, Beijing, People’s Republic of China. For the subcutaneous tumor experiments,41–43 MDA-MB-231 cells that had been infected with LV-645, control miRNA or LV-645+ uPAMut were seeded into four- to six-week old nude mice. Each nude mouse was injected with 1×106 cells. After four to six weeks of growth, the subcutaneous tumors were harvested. The tumor volume and weight were measured.
In vitro invasion or migration of MDA-MB-231 cells (transwell experiments)
MDA-MB-231 cells, which were infected with control miRNA, LV-645 (miR-645), or LV-645+ uPAMut, were harvested for transwell experiments. The in vitro invasion of MDA-MB-231 cells were measured by invasion-transwell and the in vitro migration of MDA-MB-231 cells were measured by migration-transwell following the methods provided by Liang et al33 and Li et al.34 Relative invasion cells or migration cells were measured by OD 546 nm values and inhibitory rates were calculated as ([control group OD 546 nm value − administration group OD 546 nm value]/[control group OD 546 nm value]) × 100%.
In vivo tumor growth (intrahepatic TNBC tumor model)
The animal experiments were approved by the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Xi’an Jiao Tong University. All the animal studies were conducted in accordance with the U.K. Animals (Scientific Procedures) Act of 1986 and the associated guidelines. As discussed above, MDA-MB-231 cells that had been infected with LV-645, control miRNA or LV-645+ uPAMut were seeded into four- to six-week old nude mice to prepare the subcutaneous TNBC tumor model. After the subcutaneous tumors were formed, the tumor tissues were collected in the form of microtissue samples (0.2 × 0.2 × 0.2 mm). The microtissues were adhered to the surface of the nude mice’s livers using biological-medical gel (Cai-Hong-Yi-Xue-She-Bei Corporation, Kunming City, Yunnan Province, People’s Republic of China).44,45 After four to eight weeks of growth, the mice were harvested. The liver organs with nodules formed by the MDA-MB-231 cells were collected for Masson staining, following the methods described by Shao et al and Xie et al.46,47 The photographs of the Masson staining were quantitatively analyzed to determine the relative invasive growth of the MDA-MB-231 cells using the ImageJ software, following the methods descripted by Shao et al and Xie et al.46–48 The thickness of the nodules or liver organs was determined, and the relative invasive growth of the MDA-MB-231 cells in the nude mice’s livers (the nodule thickness relative to that of the liver organ) was calculated as follows: (the nodule thickness of the control group)/(the liver organ thickness of the control group) × 100% or (the nodule thickness of the administration group)/(the liver organ thickness of the administration group) × 100%. The inhibition rate of each group was calculated as follows: ([the control group relative invasive growth) − (the administration group relative invasive growth])/(the control group relative invasive growth) × 100%.
Statistical analysis
The western blot bands were analyzed using the ImageJ software. The relative expression of the targeted proteins was calculated as follows: (the indicated group intensity of the targeted protein bands)/(the indicated group intensity of the loading control bands). The statistical analysis was performed using Bonferroni correction with or without two-way analysis of variance, SPSS software (IBM Corporation, Armonk, NY, USA). P-values <0.05 were considered statistically significant.
Results
Transduction of miR-645 decreased uPA expression by targeting the 3′ UTR of the uPA mRNA
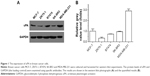
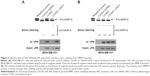
First, we examined uPA expression in several breast cancer cell lines. As shown in Figure 1, uPA expression was much higher in the MDA-MB-231 cells (an aggressive TNBC cell line) than in the MCF-7, ZR-75-1, BT474 or SK-BR3 cells (representing other types of breast cancer). Next, the effect of miR-645 on uPA expression in the MDA-MB-231 cells was examined. As shown in Figure 2A, italicized fonts indicate the potential target sequence or the mutated sequence of miR-645 within the uPA mRNA 3′ UTR. The MDA-MB-231 cells were transfected for the control miRNA or transduced for the LV particles, LV-645+ uPAMut or LV-645+, a miR-645 inhibitor for 48 hours; then the cells were harvested for western blot analysis (Figure 2B and C). LV-645 significantly decreased uPA expression compared to the control miRNA, but not compared to uPAMut, which contained a mutated miR-645-binding site (Figure 2A–C). Transfection of the miR-645 inhibitor almost blocked the effect of miR-645 on uPA expression in the MDA-MB-231 cells (Figure 2B and C). Therefore, miR-645 repressed the expression of uPA in MDA-MB-231 cells by directly targeting the uPA mRNA’s 3′ UTR.
Transduction of miR-645 inhibited the cleaving of pro-MMP-9
An in vitro biochemical assay was used to investigate the effect of miR-645 on the ability of uPA to cleave pro-MMP-9. As shown in Figure 3, the uPA protein in the MDA-MB-231 cells could be isolated and immunoprecipitated by the Protein A magnetic beads (Figure 3). The LV-645 infection significantly decreased the amount of uPA protein in the MDA-MB-231 cells (input uPA) and the amount that could be isolated by the Protein A magnetic beads (immunoprecipitated uPA). Transfection of uPAMut almost blocked the effect of LV645 on the uPA expression levels (Figure 3A). The results from the biochemical assays revealed that the uPA protein isolated by the Protein A magnetic beads directly cleaved the purified pro-MMP-9 protein (Figure 3A and B). The bands of the pro-MMP-9 or the cleaved act-MMP-9 were examined using silver staining (Figure 3A and B). Next, to examine the specificity of miR-645’s inhibitory function, the loading amount on the beads of the uPA that had been isolated by the Protein A magnetic beads was increased to make equal uPA loading amounts (amount pf uPA loaded into the gel) in the control group, the LV-645 group and the LV645+ uPAMut group (Figure 3B). As shown in Figure 3B, the amount of pro-MMP-9 cleaving was the same in all three groups. Therefore, miR-645 inhibited the expression of uPA but did not affect the enzyme activation of uPA in the MDA-MB-231 cells.
Transduction of miR-645 inhibited the subcutaneous growth of the MDA-MB-231 cells in nude mice
A subcutaneous model of MDA-MB-231 cells in nude mice was used to determine whether miR-645 could inhibit the growth of MDA-MB-231 cells. MDA-MB-231 cells that had been infected with control miRNA, LV-645 or LV-645+ uPAMut were seeded into nude mice (Figure 4). As shown in Figure 4, LV-645 inhibited the subcutaneous growth of MDA-MB-231 cells in the nude mice. Infection with uPAMut almost blocked the effect of LV-645. Therefore, miR-645 inhibited the subcutaneous growth of MDA-MB-231 cells by targeting uPA.
Transduction of miR-645 inhibited the invasive growth of the MDA-MB-231 cells in the nude mice’s liver organs
To examine whether miR-645 inhibits the invasion or migration of MDA-MB-231 cells, transwell assays were performed. As shown in Figure 5, miR-645 significantly inhibited the in vitro invasion (Figure 5A) or migration (Figure 5B). Transfection of miR-645 inhibitor or mutated uPA almost blocked the inhibitory effect of miR-645 on the in vitro invasion (Figure 5A) or migration (Figure 5B). Moreover, the inhibitory activation of miR-645 is more obvious on invasion-transwell experiments compared with migration-transwell experiments of MDA-MB-231 cells.
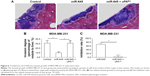
Transplantation of MDA-MB-231 cells into the nude mice’s liver organs was used as a model to mimic the in vivo invasive growth of MDA-MB-231 cells. As shown in Figure 6, the MDA-MB-231 cells broke the surface of the liver organs, destroyed the liver organ tissues and invasively grew into the liver organs. LV-645 infection significantly inhibited the invasive growth of the MDA-MB-231 cells in the liver organs. Infection with uPAMut almost blocked the inhibitory effect of LV-645. Therefore, miR-645 inhibited the in vivo invasive growth of MDA-MB-231 cells in the nude mice’s liver organs by targeting uPA.
Discussion
Invasion and migration are important factors that influence the prognosis of patients with malignant tumors.49,50 Cancer cells proliferate at the primary site to form tumor tissues, and under the combined influence of various cytokines and hypoxia in the tumor microenvironment, metastasis and invasion can occur.49,50 The ECM functions as a barrier to confine cancer cells within their primary origin site.51,52 During the invasion/migration process, ECM degradation can be induced by MMPs, a family of zinc-dependent enzymes that proteolytically degrade various ECM components; the pro-MMPs are cleaved and activated by uPA.53–56 Therefore, inhibition of uPA expression or activation is a promising approach to inhibit tumor invasion and migration.57
In the present work, miR-645 was identified using an online tool (miRDB) as potentially targeting uPA. This targeting was confirmed by western blot experiments. Transduction with LV-645 inhibited uPA expression in MDA-MB-231 cells, while transfection of an uPA expression vector with a mutated miR-645 target site or a miR-645 inhibitor almost blocked the effect of LV-645 on uPA expression. These results indicate that miR-645 suppresses uPA expression by targeting its 3′ UTR. Moreover, LV-645 infection decreased uPA expression and, in turn, decreased the uPA cleaving of pro-MMP-9. Decreasing uPA expression to attenuate the invasive growth of TNBC cells represents a promising therapeutic approach.
Breast cancer remains the leading cause of death from cancer among women.58 The prognosis of endocrine-related breast cancer and HER2-positive breast cancer has improved due to effective antitumor drugs.59,60 However, the prognosis and clinical outcomes of TNBC, which does not express estrogen receptors (ER), progesterone receptors (PR) or HER2, are far from satisfying.61,62 Although TNBC only accounts for a small portion (10%–20%) of all breast cancer cases, it has a high mortality risk and is more aggressive than other breast cancer sub-types.63 Given the absence of definitive therapeutic targets (ER, PR or HER2), only a few adjuvant treatments, such as conventional surgery, chemotherapy and radiotherapy, can provide any benefit for TNBC patients.64,65
In the present work, we used lentivirus particles to express miR-645 in MDA-MB-231 cells. Decreasing uPA expression through LV-645 significantly decreased the subcutaneous growth of the MDA-MB-231 cells. LV-645 infection also significantly decreased the invasive growth of the MDA-MB-231 cells in the nude mice’s liver organs. This intrahepatic tumor model was used to better understand the invasion process of the TNBC cells. This model is more representative of the disease than other tumor metastasis models, for example, those that involve injecting cancer cells into the tail vein to form lung tumor nodules.33,34 As the massive capillary system in the lungs allows tumor cells to migrate throughout the entire organ, the pathological invasion process of human cancer cells cannot be mimicked by this model. In our intrahepatic tumor model, microtissues formed by MDA-MB-231 cells first break through the capsule on the liver surface and then invasively proliferate throughout the liver, eventually establishing tumor nodules. This intrahepatic tumor model is a more sophisticated and suitable model to mimic the invasive growth of cancer cells in a human organ. Recently, reports of miR-645 are rare and there are no reports that mentioned the function of miR-645 in TNBC. It has been confirmed that uPA is a promising target for TNBC treatment and enhances malignant potential of TNBC cells. Therefore, targeting uPA by miR-645 is a novel approach for TNBC treatment.66–69 Moreover, MDA-MB-231 is a most typical and convince cell line for TNBC research. The other models, eg, patient-derived xenograft model, are valuable to be used.70,71
Moreover, highly aggressive growth would be the foremost feature for human malignancy tumors. The invasion or migration of cancer cells was often measured by in vitro or in vivo models. Transwell experiments often were used to examine the in vitro invasion or migration of human cancers. Results indicated that the inhibitory activation of miR-645 is more obvious in invasion-transwell experiments compared with migration-transwell experiments of MDA-MB-231 cells. This may due to the fact that miR-645 inhibits the expression of uPA, which plays central roles in degradation of ECM during invasion process compared with migration process. Our work attempted to measure the invasive growth of MDA-MB-231, a TNBC cell line with a highly aggressive feature. To establish the metastasis model of breast cancer, cells often were injected into the lateral tail vein to mimic breast cancer liver metastasis.33,34 However, the massive capillary system in the lungs would induce the location of tumor cells in the lungs, and the pathological metastasis of breast cancer cells cannot be simply mimicked by these methods. ECM degradation induced by MMPs plays central roles in invasion of human cancers, and cleaving of MMPs by uPA is essential for their activation. In the present work, MDA-MB-231 cells were initially adhered in the surface of the liver and then invade into the liver. The aggressiveness of cells in the liver is related to the degradation of ECM. Therefore, the intrahepatic tumor model that mimics the breast cancer liver metastasis is more representative compared with the regular tumor metastasis model and would be a useful tool to evaluate the therapeutic efficiency of anti-tumor therapies. However, this model is highly suitable to mimic the invasive growth of breast cancer, but not easily to investigate the extravasation of cells from vessels to the secondary organs. There is an urgent need to establish a model that not only mimics the invasive growth of cancer cells but also reflects the extravasation of cancer cells from vessels to the secondary organs.
MiR is an important component of non-coding RNA molecules and the epigenetic regulation of human gene expression.72 In the present work, we demonstrated that miR-645 targets uPA. We used miR-645 lentivirus particles to repress uPA expression and the invasive growth of MDA-MB-231 cells. Other miRs, such as miR-193b or RNA-23b, could also function as uPA regulators and participate in the regulation of human cancer metastasis.73–77 Future studies should focus on the tissue specificity of these miRs for potential application in treating human cancers with diverse tissue origins.
Conclusion
This study demonstrated that uPA is targeted by miR-645. Infection of miR-645 lentivirus particles suppressed uPA expression and inhibits the invasive growth of MDA-MB-231 TNBC cells. Downregulation of uPA by miR-645 represents a promising strategy for TNBC treatment.
Author contributions
All the authors made substantial contributions to the design and conception, acquisition, analysis or interpretation of data. The authors took part in either drafting or revising the manuscript. At the same time, the authors gave final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
Dasgupta S, Rajapakshe K, Zhu B, et al. Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer. Nature. 2018;556(7700):249–254. | ||
Lyman GH, Greenlee H, Bohlke K, et al. Integrative therapies during and after breast cancer treatment: ASCO Endorsement of the SIO Clinical Practice Guideline. J Clin Oncol. 2018;36(25):2647–2655. | ||
Slamon DJ, Neven P, Chia S, et al. Phase III randomized study of ribociclib and fulvestrant in hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: MONALEESA-3. J Clin Oncol. 2018;36(24):2465–2472. | ||
Li J, Li B, Jiang Q, et al. Do genetic polymorphisms of the vitamin D receptor contribute to breast/ovarian cancer? A systematic review and network meta-analysis. Gene. 2018;677:211–227. | ||
Sachs N, de Ligt J, Kopper O, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1–2):373.e10–386.e10. | ||
Xiong Z, Deng G, Huang X, et al. Bone metastasis pattern in initial metastatic breast cancer: a population-based study. Cancer Manag Res. 2018;10:287–295. | ||
Zhang F, Linghu R, Zhan X, et al. Efficacy, safety and proper dose analysis of PEGylated granulocyte colony-stimulating factor as support for dose-dense adjuvant chemotherapy in node positive Chinese breast cancer patients. Oncotarget. 2017;8(45):80020–80028. | ||
Zhang F, Yang J, Li H, et al. Combating rituximab resistance by inducing ceramide/lysosome-involved cell death through initiation of CD20-TNFR1 co-localization. Oncoimmunology. 2016;5(5):e1143995. | ||
Zhang F, Zhang J, Liu M, et al. Combating HER2-overexpressing breast cancer through induction of calreticulin exposure by Tras-Permut CrossMab. Oncoimmunology. 2015;4(3):e994391. | ||
Fasching PA, Loibl S, Hu C, et al. BRCA1/2 Mutations and bevacizumab in the neoadjuvant treatment of breast cancer: response and prognosis results in patients with triple-negative breast cancer from the GeparQuinto study. J Clin Oncol. 2018;36(22):2281–2287. | ||
Keren L, Bosse M, Marquez D, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174(6):e19:1373–1387. | ||
Shah N, Mohammad AS, Saralkar P, et al. Investigational chemotherapy and novel pharmacokinetic mechanisms for the treatment of breast cancer brain metastases. Pharmacol Res. 2018;132:47–68. | ||
Zhu Z, Li Y, Yang X, Pan W, Pan H. The reversion of anti-cancer drug antagonism of tamoxifen and docetaxel by the hyaluronic acid-decorated polymeric nanoparticles. Pharmacol Res. 2017;126:84–96. | ||
Williams MM, Lee L, Werfel T, et al. Intrinsic apoptotic pathway activation increases response to anti-estrogens in luminal breast cancers. Cell Death Dis. 2018;9(2):21. | ||
Zhang J, Zhou C, Jiang H, et al. ZEB1 induces ER-α promoter hypermethylation and confers antiestrogen resistance in breast cancer. Cell Death Dis. 2017;8(4):e2732. | ||
Aghazadeh S, Yazdanparast R. Activation of STAT3/HIF-1α/Hes-1 axis promotes trastuzumab resistance in HER2-overexpressing breast cancer cells via down-regulation of PTEN. Biochim Biophys Acta. 1861;2017:1970–1980. | ||
Centelles MN, Wright M, Gedroyc W, Thanou M. Focused ultrasound induced hyperthermia accelerates and increases the uptake of anti-HER-2 antibodies in a xenograft model. Pharmacol Res. 2016;114:144–151. | ||
Ouchida AT, Li Y, Geng J, et al. Synergistic effect of a novel autophagy inhibitor and Quizartinib enhances cancer cell death. Cell Death Dis. 2018;9(2):138. | ||
Aydinlik S, Erkisa M, Cevatemre B, et al. Enhanced cytotoxic activity of doxorubicin through the inhibition of autophagy in triple negative breast cancer cell line. Biochim Biophys Acta Gen Subj. 2017;1861(2):49–57. | ||
Harris NLE, Vennin C, Conway JRW. Australian Pancreatic Cancer Genome Initiative, Biankin AV, Ranson M, Timpson P, Saunders DN. SerpinB2 regulates stromal remodelling and local invasion in pancreatic cancer. Oncogene. 2017;36:4288–4298. | ||
Kim MJ, Kim WS, Kim DO, et al. Macrophage migration inhibitory factor interacts with thioredoxin-interacting protein and induces NF-κB activity. Cell Signal. 2017;34:110–120. | ||
Ghasemi A, Hashemy SI, Aghaei M, Panjehpour M. RhoA/ROCK pathway mediates leptin-induced uPA expression to promote cell invasion in ovarian cancer cells. Cell Signal. 2017;32:104–114. | ||
Paez AV, Pallavicini C, Schuster F, et al. Heme oxygenase-1 in the forefront of a multi-molecular network that governs cell-cell contacts and filopodia-induced zippering in prostate cancer. Cell Death Dis. 2016;7(12):e2570. | ||
Pavet V, Shlyakhtina Y, He T, et al. Plasminogen activator urokinase expression reveals TRAIL responsiveness and supports fractional survival of cancer cells. Cell Death Dis. 2014;5:e1043. | ||
Dinesh P, Rasool M. uPA/uPAR signaling in rheumatoid arthritis: Shedding light on its mechanism of action. Pharmacol Res. 2018;134:31–39. | ||
Li S, Wei X, He J, Tian X, Yuan S, Sun L. Plasminogen activator inhibitor-1 in cancer research. Biomed Pharmacother. 2018;105:83–94. | ||
Seeley JJ, Baker RG, Mohamed G, et al. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature. 2018;559(7712):114–119. | ||
Li Y, Cui X, Li Y, Zhang T, Li S. Upregulated expression of miR-421 is associated with poor prognosis in non-small-cell lung cancer. Cancer Manag Res. 2018;10:2627–2633. | ||
Duan F, Yang Y, Liu W, et al. Quantifying the prognostic significance of microRNA-17/17-5P in cancers: a meta-analysis based on published studies. Cancer Manag Res. 2018;10:2055–2069. | ||
Sen K, Bhattacharyya D, Sarkar A, et al. Exploring the major cross-talking edges of competitive endogenous RNA networks in human Chronic and Acute Myeloid Leukemia. Biochim Biophys Acta Gen Subj. 2018;1862(9):1883–1892. | ||
Li L, Liang Y, Kang L, et al. Transcriptional regulation of the warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3):368–385. | ||
Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8(10):e3103. | ||
Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8(7):e2928. | ||
Li L, Kang L, Zhao W, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. | ||
Yang Q, Feng F, Zhang F, et al. LINE-1 ORF-1p functions as a novel HGF/ETS-1 signaling pathway co-activator and promotes the growth of MDA-MB-231 cell. Cell Signal. 2013;25(12):2652–2660. | ||
Zhang F, Feng F, Yang P, et al. Four-and-a-half-LIM protein 1 down-regulates estrogen receptor α activity through repression of AKT phosphorylation in human breast cancer cell. Int J Biochem Cell Biol. 2012;44(2):320–326. | ||
Zhu M, Li M, Zhang F, et al. FBI-1 enhances ETS-1 signaling activity and promotes proliferation of human colorectal carcinoma cells. PLoS One. 2014;9(5):e98041. | ||
Lu Y, Feng F, Yang Y, et al. LINE-1 ORF-1p functions as a novel androgen receptor co-activator and promotes the growth of human prostatic carcinoma cells. Cell Signal. 2013;25(2):479–489. | ||
Zhao Y, Lyons CE, Xiao A, et al. Urokinase directly activates matrix metalloproteinases-9: a potential role in glioblastoma invasion. Biochem Biophys Res Commun. 2008;369(4):1215–1220. | ||
Wei S, Chen Y, Chung L, Nagase H, Brew K. Protein engineering of the tissue inhibitor of metalloproteinase 1 (TIMP-1) inhibitory domain. In search of selective matrix metalloproteinase inhibitors. J Biol Chem. 2003;278(11):9831–9834. | ||
An L, Li DD, Chu HX, et al. Terfenadine combined with epirubicin impedes the chemo-resistant human non-small cell lung cancer both in vitro and in vivo through EMT and Notch reversal. Pharmacol Res. 2017;124:105–115. | ||
Chen Z, Jiang Z, Zhang W, He B, Bx H. Silencing the expression of copine-III enhances the sensitivity of hepatocellular carcinoma cells to the molecular targeted agent sorafenib. Cancer Manag Res. 2018;10:3057–3067. | ||
Jia H, Yang Q, Wang T, et al. Rhamnetin induces sensitization of hepatocellular carcinoma cells to a small molecular kinase inhibitor or chemotherapeutic agents. Biochim Biophys Acta. 2016;1860(7):1417–1430. | ||
Feng F, Jiang Q, Cao S, et al. Pregnane X receptor mediates sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta Gen Subj. 2018;1862(4):1017–1030. | ||
Zhang Y, Li D, Jiang Q, et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of Sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9(7):743. | ||
Shao Z, Li Y, Dai W, et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188–200. | ||
Xie H, Tian S, Yu H, et al. A new apatinib microcrystal formulation enhances the effect of radiofrequency ablation treatment on hepatocellular carcinoma. Onco Targets Ther. 2018;11:3257–3265. | ||
Xie H, Yu H, Tian S, et al. MEIS-1 level in unresectable hepatocellular carcinoma can predict the post-treatment outcomes of radiofrequency ablation. Oncotarget. 2018;9(20):15252–15265. | ||
Xu J, Wang F, Wang X, He Z, Zhu X. miRNA-543 promotes cell migration and invasion by targeting SPOP in gastric cancer. Onco Targets Ther. 2018;11:5075–5082. | ||
Chen X, Qu J. Long non-coding RNA MEG3 suppresses survival, migration, and invasion of cervical cancer. Onco Targets Ther. 2018;11:4999–5007. | ||
Stylianou A, Gkretsi V, Stylianopoulos T. Transforming growth factor-β modulates pancreatic cancer associated fibroblasts cell shape, stiffness and invasion. Biochim Biophys Acta. 1862;2018:1537–1546. | ||
Singh R, Dagar P, Pal S, Basu B, Shankar BS. Significant alterations of the novel 15 gene signature identified from macrophage-tumor interactions in breast cancer. Biochim Biophys Acta Gen Subj. 2018;1862(3):669–683. | ||
Zaremba-Czogalla M, Hryniewicz-Jankowska A, Tabola R, et al. A novel regulatory function of CDKN1A/p21 in TNFα-induced matrix metalloproteinase 9-dependent migration and invasion of triple-negative breast cancer cells. Cell Signal. 2018;47:27–36. | ||
Kim MJ, Kim WS, Kim DO, et al. Macrophage migration inhibitory factor interacts with thioredoxin-interacting protein and induces NF-κB activity. Cell Signal. 2017;34:110–120. | ||
Uemura T, Suzuki T, Saiki R, et al. Activation of MMP-9 activity by acrolein in saliva from patients with primary Sjögren’s syndrome and its mechanism. Int J Biochem Cell Biol. 2017;88:84–91. | ||
Lee SY, Na YJ, Jeong YA, Kim JL, Oh SC, Lee DH. Upregulation of EphB3 in gastric cancer with acquired resistance to a FGFR inhibitor. Int J Biochem Cell Biol. 2018;102:128–137. | ||
Völker HU, Weigel M, Strehl A, Frey L. Levels of uPA and PAI-1 in breast cancer and its correlation to Ki67-index and results of a 21-multigene-array. Diagn Pathol. 2018;13(1):67. | ||
Zhang K, Hong R, Xu F, et al. Clinical value of circulating ESR1 mutations for patients with metastatic breast cancer: a meta-analysis. Cancer Manag Res. 2018;10:2573–2580. | ||
Gu S, Liang H, Qi D, et al. Knockdown of KIF26B inhibits breast cancer cell proliferation, migration, and invasion. Onco Targets Ther. 2018;11:3195–3203. | ||
Xie W, Zhang Y, He Y, et al. A novel recombinant human Frizzled-7 protein exhibits anti-tumor activity against triple negative breast cancer via abating Wnt/β-catenin pathway. Int J Biochem Cell Biol. 2018;103:45–55. | ||
Sulaiman A, Mcgarry S, Lam KM, et al. Co-inhibition of mTORC1, HDAC and ESR1α retards the growth of triple-negative breast cancer and suppresses cancer stem cells. Cell Death Dis. 2018;9(8):815. | ||
Wang S, Ke H, Zhang H, et al. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Dis. 2018;9(8):805. | ||
Yuan N, Zhang G, Bie F, et al. Integrative analysis of lncRNAs and miRNAs with coding RNAs associated with ceRNA crosstalk network in triple negative breast cancer. Onco Targets Ther. 2017;10:5883–5897. | ||
Denard B, Jiang S, Peng Y, Ye J. CREB3L1 as a potential biomarker predicting response of triple negative breast cancer to doxorubicin-based chemotherapy. BMC Cancer. 2018;18(1):813. | ||
Li J, Lai Y, Ma J, et al. miR-17-5p suppresses cell proliferation and invasion by targeting ETV1 in triple-negative breast cancer. BMC Cancer. 2017;17(1):745. | ||
Huber MC, Mall R, Braselmann H, et al. uPAR enhances malignant potential of triple-negative breast cancer by directly interacting with uPA and IGF1R. BMC Cancer. 2016;16:615. | ||
Li X, Wu B, Chen L, Ju Y, Li C, Meng S. Urokinase-type plasminogen activator receptor inhibits apoptosis in triple-negative breast cancer through miR-17/20a suppression of death receptors 4 and 5. Oncotarget. 2017;8(51):88645–88657. | ||
Woodcock CC, Huang Y, Woodcock SR, et al. Nitro-fatty acid inhibition of triple-negative breast cancer cell viability, migration, invasion, and tumor growth. J Biol Chem. 2018;293(4):1120–1137. | ||
Szarc Vel Szic K, Declerck K, Crans RAJ, et al. Epigenetic silencing of triple negative breast cancer hallmarks by Withaferin A. Oncotarget. 2017;8(25):40434–40453. | ||
Rosato RR, Dávila-González D, Choi DS, et al. Evaluation of anti-PD-1-based therapy against triple-negative breast cancer patient-derived xenograft tumors engrafted in humanized mouse models. Breast Cancer Res. 2018;20(1):108. | ||
Eckhardt BL, Gagliardi M, Iles L, et al. Clinically relevant inflammatory breast cancer patient-derived xenograft-derived ex vivo model for evaluation of tumor-specific therapies. PLoS One. 2018;13(5):e0195932. | ||
Hines PJ. MicroRNAs in functional and dysfunctional pain. Science. 2017;356(6343):1134–1136. | ||
Iwamoto N, Vettori S, Maurer B, et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann Rheum Dis. 2016;75(1):303–310. | ||
Li J, Kong F, Wu K, Song K, He J, Sun W. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol Med Rep. 2014;10(5):2613–2620. | ||
Xie C, Jiang XH, Zhang JT, et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32(18):2291.e1–7:2282–2291. | ||
Li XF, Yan PJ, Shao ZM. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene. 2009;28(44):3937–3948. | ||
Salvi A, Sabelli C, Moncini S, et al. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. Febs J. 2009;276(11):2966–2982. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.