Back to Journals » Neuropsychiatric Disease and Treatment » Volume 14
microRNA-451 protects neurons against ischemia/reperfusion injury-induced cell death by targeting CELF2
Authors Liu Q, Hu Y , Zhang M, Yan Y, Yu H, Ge L
Received 9 May 2018
Accepted for publication 28 July 2018
Published 23 October 2018 Volume 2018:14 Pages 2773—2782
DOI https://doi.org/10.2147/NDT.S173632
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Yuping Ning
Qian Liu,1 Yaguang Hu,2 Min Zhang,3 Yousheng Yan,4 Hongmei Yu,1 Li Ge1
1Department of Neurological Rehabilitation, Gansu Provincial Hospital, Gansu 730000, China; 2Department of Pharmacy, Gansu Provincial Maternity and Child-Care Hospital, Gansu 730050, China; 3Department of Pathology, Gansu Provincial Hospital, Gansu 730000, China; 4National Centre for Human Genetic Resources, National Research Institute for Health and Family Planning, Beijing 100089, China
Objectives: miRNAs are a family of non-coding RNAs that affect cell growth, migration and apoptosis. However, little is known on the behavior of miRNAs in neurons. Hence, this work aimed to investigate the functions and roles of miRNA-451 in neurons induced by ischemia/reperfusion injury.
Materials and methods: In this study, we established a 12- or 24-hour oxygen and glucose deprivation/reoxygenation (OGD/R) cell model. miR-451 mimic, si-CUGBP Elav-like family member 2 (siCELF2), oeCELF2 and the corresponding negative controls were transfected into the 24-hour OGD/R cells. The transfection efficiency and the relative expression of miR-451 and CELF2 were measured using quantitative reverse transcription PCR and Western blot analysis. Cell viability, apoptosis, oxidative stress and cleaved-caspase-3 expression were assessed using Cell Counting Kit-8, LDH, SOD, malondialdehyde, ROS assays, flow cytometry and Western blot analysis upon miR-451 overexpression, CELF2 silencing or overexpression of both. Bioinformatics analysis and the dual-luciferase reporter assay were used to examine the relationship between CELF2 and miR-451 in the OGD/R cells.
Results: The results showed that miR-451 was downregulated in the OGD/R cells. The overexpression of miR-451 increased cell viability and SOD activity, but decreased apoptosis rate, levels of LDH, MDA, ROS and cleaved caspase-3 expression. CELF2 silencing inhibited apoptosis and oxidative stress. The results suggested that CELF2 was a target of miR-451, and that CELF2 overexpression alleviated the inhibitory effect of miR-451 on apoptosis and oxidative stress of the OGD/R cells.
Conclusion: The results demonstrated that miR-451 could protect cells against OGD/R-induced apoptosis and oxidative stress by targeting CELF2.
Keywords: miR-451, CELF2, I/R injury, neuron, oxidative stress, apoptosis
Introduction
Ischemic peripheral neuropathy is an occlusive vascular disease.1 Neurological diseases, such as cerebral arterial thrombosis, Alzheimer’s disease and Parkinson’s disease, have become a major concern of human health worldwide.2 A series of endogenous injuries are caused when blood is restored and perfused posterior to the ischemia, that is, the cerebral ischemia/reperfusion (I/R) injury.3,4 During this process, a large number of harmful factors such as reactive oxygen species are released to the vascular micro-environment, damaging the cell membrane structure and mitochondrial function and ultimately leading to apoptosis.3,5,6 Scavenging oxygen-free radicals to inhibit such an apoptosis is, therefore, critical in the protection of neurons induced by I/R injury.
miRNAs are a class of non-coding RNAs with a length of 22 nucleotides.7 They bind specifically to the 3′-UTR of the target gene to degrade or inhibit the translation of target mRNA, regulating the gene post-transcriptionally.8,9 Previous studies on miRNAs mainly focused on the occurrence of tumors and immune diseases;10 recent studies have found that miRNAs are also involved in the pathogenesis of many central nervous system tumors and neurodegenerative diseases.11,12 miRNAs are closely related to ischemic neurological diseases.2 Jeyaseelan et al reported that 106 miRNAs were found abnormally expressed in the rats middle cerebral artery occlusion model.13 Ji et al showed that miR-9 and miR-124 could be used as biomarkers to assess the degree of injury caused by I/R in acute ischemic stroke.14 Although miRNAs are closely related to I/R injury, only a few miRNAs have been proved to be associated with specific diseases with their regulatory mechanisms remaining unknown.
miR-451 is located on chromosome 17q11.2, adjacent to the proto-oncogene human epidermal growth factor receptor 2.15,16 Its abnormal expression has been recently detected in breast, colorectal and lung cancers.17–19 However, little is known on the relationship between miR-451 and ischemic neuropathy and the corresponding regulatory mechanism.
CELF2 is a RNA-binding protein and is widely expressed in developmental and adult tissues of organisms.20 It could regulate alternative splicing of pre-mRNA and might play an important role in mRNA editing, shuttling and translation.21 It was reported that the CELF2-encoding gene was located on chromosome 10, the deletion of which was related to the occurrence and development of glioma.22 Furthermore, a tissue-specific subtype of CELF2 was found in eyes, heart, liver and nervous system.20 However, little is known about CELF2 in neural disease. Miyazaki et al reported that miR-196a enhanced the decay of androgen receptor mRNA through silencing CELF2 to inhibit spinal and bulbar muscular atrophy.23
In this work, the regulatory mechanism and the effects of miR-451 on I/R-damaged neurons in ischemic neuropathy, as well as the potential relationship between miR-451 and CELF2 were investigated.
Materials and methods
Cell culture and the establishment of the oxygen and glucose deprivation/reoxygenation (OGD/R) cells model
The PC-12 cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM/Nutrient F-12 Ham (DMEM/F12; Sigma-Aldrich Co., St Louis, MO, USA) supplemented with 10% (v/v) FBS (Sigma-Aldrich Co.). In order to establish an OGD/R cell model, the cells were seeded in 96-well plates (5×103 cells/well) at 37°C for 24 hours. The supernatant was removed, and the growth medium was replaced with the glucose-free and serum-free DMEM. Cells were then leftsupp in the hypoxic chambers with a supply of 95% N2 and 5% CO2 for 20 minutes at a rate of 20 L/min. Subsequently, the outlet valve was clamped, and the chambers were incubated in a humidified incubator at 37°C for 12 or 24 hours. Oxygen supply was recovered after hypoxia OGD experiment and the cells were incubated at 37°C for 12 hours in a humidified atmosphere containing 5% CO2.
Cell transfection
The OGD/R cells were seeded in 12-well plates (1×105 cells/well) at 37°C. The cells were digested upon 60%–70% confluence using 0.25% pancreatin (Sigma-Aldrich Co.). miR-451 mimic (5′-AAACCGUUACCAUUACUGAGUU-3′) and its negative control (NC; 5′-UCGCUUGGUGCAGGUCGGGAA-3′) were purchased from GenePharma (Shanghai, China), and Lipofectamine 3000 Reagent (Thermo Fisher Scientific, Waltham, MA, USA) was mixed and diluted at room temperature for 6 hours. siRNA CELF2 (siCELF2), oeCELF2 (for CELF2 silencing expression and overexpression, respectively) and their corresponding NCs were purchased from GenePharma. High-glucose DMEM medium and the Lipo6000™ Transfection Reagent (Beyotime, Shanghai, China) were mixed for 20 minutes. The sequences were as follows: 5′-CACCTATCGTGGTGAAGTT-3′, 5′-CACAGTATCTGGCGCTCCT-3′ and 5′-GAATGCACTGCACAATATT-3′. All cells were replaced after 5–6 hours of incubation and collected after 48 hours for further experiments.
Cell viability assay
Cells were seeded into 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. The Cell Counting Kit-8 (Beyotime) was used to detect cell viability. The pre-made Cell Counting Kit-8 solution (10 μL/well) was added to the cell culture (200 μL), which was then incubated at 37°C for 1 hour in a humidified atmosphere containing 5% CO2. The absorbance at 450 nm was measured using Microplate Reader (Bio-Rad, Hercules, CA, USA).
Cytotoxicity assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. Cytotoxicity was measured using the LDH Cytotoxicity Assay Kit (Beyotime). LDH release regent (150 μL) was added to the cell culture plate with the supernatant removed by centrifugation. The cells were incubated at 37°C for 1 hour in a humidified atmosphere containing 5% CO2. The absorbance at 490 nm was measured using Microplate Reader (Bio-Rad).
SOD assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. SOD assay was performed using the Total Superoxide Dismutase Assay Kit with WST-8 (Beyotime). Cells were washed with cold PBS, followed by the addition of SOD buffer solution. Protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime). The samples (20 μL) was mixed with the WST-8/enzyme solution (160 μL) and the reaction starting solution (20 μL) and then incubated at 37°C for 30 minutes. The absorbance at 450 nm was measured using Microplate Reader (Bio-Rad).
Malondialdehyde (MDA) assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. The MDA assay was performed using the Lipid Peroxidation MDA Assay Kit (Beyotime). Cells were lysed by cell lysis buffer (Beyotime) and the protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime). The samples (0.1 mL) was mixed with the MDA solution (0.2 mL) and heated for 15 minutes. The absorbance at 532 nm was measured using Microplate Reader (Bio-Rad) after the samples were cooled to room temperature.
ROS assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. The ROS assay was performed using the Reactive Oxygen Species Assay Kit (Beyotime). Cells were collected, suspended in the dichlorofluorescin diacetate solution (1,000× diluted) and incubated at 37°C for 20 minutes. Cells were washed three times with cell culture. The ROS were stimulated up for 20 minutes. The cells were then observed under a fluorescence microscope (Thermo Fisher Scientific), and the ROS expression was measured using Microplate Reader (Bio-Rad).
Apoptosis assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. Cell apoptosis analysis was performed using the Annexin V-fluorescein isothiocyanate Apoptosis Detection Kit (Beyotime). Cells were washed with PBS for three times, stained in the binding buffer (200 μL) containing fluorescein isothiocyanate-Annexin V (10 μL) in the presence RNase A (50 μg/mL) and then incubated at room temperature for 1 hour in the dark. FACScan (Beckman Coulter, Fullerton, CA, USA) was used to detect differentiation of apoptotic cells (Annexin-V positive).
Dual-luciferase reporter assay
Cells were seeded in 96-well plates (5×103 cells/well) and incubated at 37°C for 48 hours. miR-451 mimics and NC mimics were co-transfected with the reporter vector CELF2 wild-type (3′UTR-WT) or CELF2 mutated-type (3′UTR-MUT) into the OGD/R cells. Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System Protocol (Promega Corporation, Fitchburg, WI, USA).
Quantitative reverse transcription PCR (qRT-PCR)
The total RNA of the PC-12 and the OGD/R cells was extracted using the Trizol Reagent (Thermo Fisher Scientific). The Taqman MicroRNA Reverse Transcription Kit (Sigma-Aldrich Co.) and Taqman Universal Master Mix II (Sigma-Aldrich Co.) were used for the qRT-PCR analysis. GAPDH was used for normalizing CELF2 and miR-451 levels. The PCR primer sequences of miR-451, CELF2 and GAPDH are shown in Table 1. Fold changes were calculated by using the relative quantification (2−ΔΔCt) method.24
  | Table 1 The sequence of the primers for qRT-PCR |
Western blot analysis
The protein was extracted using the RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific), separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membrane. The antibodies of cleaved-caspase-3 and CELF2 (1:1,000; Abcam, Shanghai, China) were prepared in a 5% blocking buffer, incubated with the membrane at 4°C overnight and then cultivated with the appropriate secondary antibody (1:2,000) marked by horseradish peroxidase at room temperature for 2 hours. The blots and antibodies were processed by the Bio-Rad ChemiDoc™ XRS system, followed by the addition of the Immobilon Western Chemiluminescent HRP Substrate (200 μL; EMD Millipore, Billerica, MA, USA). The signals were captured using the Image Lab™ Software (Bio-Rad).
Statistical analysis
The results were reported as the mean±SD. Statistical analyses were performed using the GraphPad Prism 6.0 statistical software. The P-values were calculated using the Student’s t-test or one-way ANOVA. A P-value of <0.05 was considered to indicate a statistically significant result.
Results
miR-451 expression was decreased in OGD/R cells
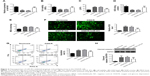
To assess the expression of miR-451 in hypoxia cells, OGD/R cell models were established and the mRNA level of miR-451 was measured using qRT-PCR. As shown in Figure 1, miR-451 expression in the OGD/R 12- and 24-hour groups was significantly lower than that in the normoxia group, with the 24-hour group being the lowest (P<0.01 or P<0.001). The 24-hour OGD/R cell model was thus chosen for the following experiments.
Overexpression of miR-451 inhibited apoptosis and oxidative stress in OGD/R cells
In order to study the effects of miR-451 on OGD/R cells, cell viability, apoptosis, cytotoxicity and oxidative stress were measured in this study. The qRT-PCR results showed that miR-451 mimic had been successfully transfected into the OGD/R cells (P<0.001; Figure 2A). It was found that cell viability and SOD activity were significantly reduced in the OGD/R cells compared to those in the normoxia cells, and that miR-451 overexpression notably alleviated such decreases (P<0.001; Figure 2B and D). On the contrary, the levels of LDH, MDA, ROS and apoptosis significantly increased when the cells were under OGD/R conditions. The observed increases were reversed posterior to the transfection of miR-451 mimic into the OGD/R cells (P<0.001; Figure 2C and E–G). As shown in Figure 2H, the results of Western blot indicated an increase in the phosphorylation level of cleaved-caspase-3 expression in the OGD/R group, but a decrease in the OGD/R+miR-451 group (P<0.001), compared with their controls. To sum up, miR-451 overexpression increased cell viability and reduced apoptosis and oxidative stress in the OGD/R cells.
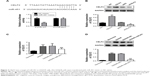
CELF2 was a target of miR-451
A previous study reported that miR-95-3 p suppression could inhibit cell proliferation by targeting CELF2 in glioma cells.22 CELF2 was selected as a potential target using the TargetScan (http://www.targetscan.org/). The OGD/R cells were transfected with miR-451 mimic or NC mimic to investigate the relationship between miR-451 expression and CELF2. In Figure 3A, the potential binding sequences between miR-451 and CELF2 were examined by bioinformatics analysis. Luciferase activity remarkably decreased upon miR-451 overexpression in the OGD/R cells transfected with CELF2-WT (P<0.001), but remained the same in the OGD/R cells transfected with CELF2-MUT. Silencing efficiency and overexpression efficiency of CELF2 are shown in Figure 3B. Expression levels of mRNA and protein of CELF2 were measured by using qRT-PCR and Western blot analysis. The results suggested that the expression of CELF2 was notably upregulated in the OGD/R cells, while miR-451 overexpression significantly inhibited such an upregulation (P<0.001; Figure 3C and D).
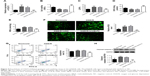
Knockdown of CELF2 inhibited apoptosis and oxidative stress in OGD/R cells
The influence of CELF2 on the OGD/R cells was subsequently investigated. As shown in Figure 4A, the expression of CELF2 increased in the OGD/R cells, but significantly decreased in the OGD/R+siCELF2 group, compared with the OGD/R+NC group (P<0.001). The data suggested that CELF2 was transfected into cells successfully with high transfection efficiency; cell viability and SOD activity remarkably increased upon CELF2 silencing in the OGD/R cells (P<0.001; Figure 4B and D). Conversely, the levels of LDH, MDA, ROS, phosphorylation of cleaved-caspase-3 expression and apoptosis significantly decreased in the OGD/R+siCELF2 group, compared with the OGD/R+NC group (P<0.001; Figure 4C and E–H). The results suggested that CELF2 silencing promoted cell viability and inhibited apoptosis and oxidative stress in the OGD/R cells.
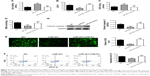
CELF2 overexpression abolished the inhibitory effect of miR-451 on apoptosis and oxidative stress of OGD/R cells
The relationship between the expression of miR-451 and CELF2 was further studied by assessing the influences of miR-451 and CELF2 overexpression on the OGD/R cells. In Figure 5A and C, it is found that miR-451 overexpression increased cell viability and SOD activity; however, they were both reduced when miR-451 and CELF2 were co-transfected into the OGD/R cells (P<0.001). As shown in Figure 5B and D–G, the levels of LDH, MDA, ROS, expression of cleaved-caspase-3 and apoptosis rate decreased in the OGD/R cells transfected with miR-451. CELF2 overexpression removed the inhibitory effects (P<0.001). The results suggested that miR-451 reduced apoptosis and oxidative stress in the OGD/R cells through CELF2 knockdown.
Discussion
Previous studies showed that scavenging oxygen-free radicals and inhibiting apoptosis were critical in terms of I/R-injured neurons’ protection. In this study, it was found that the expression of miR-451 was downregulated in OGD/R cells. miR-451 overexpression was found to be able to increase cell viability and decrease apoptosis and oxidative stress. Similarly, knockdown of CELF2 inhibited apoptosis and oxidative stress. The results also suggested that CELF2 was a target of miR-451 in the OGD/R cells. In addition, CELF2 overexpression removed the inhibitory effect of miR-451 on apoptosis and oxidative stress of the OGD/R cells.
Godlewski et al reported that miR-451 levels were regulated by glucose.25 The expression level of miR-451 was upregulated and cell proliferation was promoted upon glucose abundance in glioma cells. In contrast, miR-451 expression was downregulated by glucose deficiency, thereby decelerating cell proliferation.25 Hence, the OGD/R cell models were established in vitro to mimic I/R injury in this work. The results showed that miR-451 expression was downregulated in the OGD/R cells, consistent with previous studies. Past studies showed that miR-451 overexpression might inhibit cell growth, proliferation and migration in vitro and in vivo, accelerating the apoptosis and increasing the sensitivity of cell radiotherapy in several cancers.17–19 It was also reported that upregulation of miR-451 protected cells against cardiomyocyte anoxia/reoxygenation and alleviated loss of cardiomyocyte viability.26,27 However, our experimental results showed the opposite: miR-451 overexpression increased OGD/R cell viability and decreased apoptosis.
LDH is an enzyme that is present in the cytoplasm and can be released outside the cells upon cell damage or cell death. Its release is often used as an indicator of cytotoxicity.28 MDA is a byproduct of cell membrane lipid oxidation by ROS and a biomarker of oxidative stress of cells.29 SOD is the main antioxidant enzyme in cells playing a role in scavenging oxygen-free radicals. It can resist the damage of oxygen-free radicals to cells.30 Therefore, in this study, the levels of LDH, MDA, SOD activity and ROS in the OGD/R cells were examined. The results suggested that miR-451 increased SOD activity, but decreased the levels of LDH, MDA and ROS. The data indicated that miR-451 could scavenge oxidative stress products produced by the I/R injury and increase the activity of antioxidant enzymes.
Considering the fact that one miRNA can regulate multiple mRNA genes and different miRNAs can co-regulate one mRNA gene, a complex regulatory network is formed between miRNAs and their target genes, which controls the vital activities of cells and individuals.8 CELF2 is the basis of conservative, developmental regulation and tissue-specific processes in vertebrate embryos. Different tissues may have unique CELF2-mediated functions of nucleus and cytoplasm.20 Previous studies suggested that CELF2 was a potential tumor suppressor and played a crucial role in tumor initiation.31 Its expression could also affect the changes of splicing and development of thymic T-cells.32 Fan et al reported that miR-95-3 p suppression promoted apoptosis by targeting CELF2 in glioma cells.22 Miyazaki et al found that CELF2 directly acted on AR mRNA to reduce the total amount of abnormal androgen receptor proteins, the accumulation of which was the cause of spinal and bulbar muscular atrophy (neurodegenerative disease).23 However, the role of CELF2 in I/R disease was seldom studied. We, therefore, used TargetScan to predict the target genes of miR-451 and found that there was a potential relationship between CELF2 and miR-451. The binding sites in the 3′UTR region of CELF2 for miR-451 were then confirmed by using the dual-luciferase reporter assay, suggesting that CELF2 was a target of miR-451 in the OGD/R cells. It was also found that CELF2 overexpression could promote apoptosis and oxidative stress and remove the inhibitory effect of miR-451 on apoptosis and oxidative stress of the OGD/R cells.
In conclusion, the results above demonstrated that miRNA-451 could protect the cells against I/R-induced apoptosis and oxidative stress by targeting CELF2. This study provided a theoretical basis for further investigations on the protection of neurons against I/R injury. Other target post-I/R injury miRNAs for neurological diseases will be explored in future studies.
Disclosure
The authors report no conflicts of interest in this work.
References
Kim YA, Kim ES, Hwang HK, et al. Prevalence and risk factors for the peripheral neuropathy in patients with peripheral arterial occlusive disease. Vasc Specialist Int. 2014;30(4):125–132. | ||
Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle”. J Neuropathol Exp Neurol. 2007;66(6):461–468. | ||
Wang TF, Lei Z, Li YX, et al. Oxysophoridine protects against focal cerebral ischemic injury by inhibiting oxidative stress and apoptosis in mice. Neurochem Res. 2013;38(11):2408–2417. | ||
Lu YT, Liu SF, Mitchell JA, Malik AB, Hellewell PG, Evans TW. The role of endogenous nitric oxide in modulating ischemia–reperfusion injury in the isolated, blood-perfused rat lung. Am J Respir Crit Care Med. 1998;157(1):273–279. | ||
Li J, Tang B, Qu Y, Mu D. Telomerase reverse transcriptase: a novel neuroprotective mechanism involved in neonatal hypoxic-ischemic brain injury. Int J Dev Neurosci. 2011;29(8):867–872. | ||
Panickar KS, Anderson RA. Effect of polyphenols on oxidative stress and mitochondrial dysfunction in neuronal death and brain edema in cerebral ischemia. Int J Mol Sci. 2011;12(11):8181–8207. | ||
Li W, Ruan K. MicroRNA detection by microarray. Anal Bioanal Chem. 2009;394(4):1117–1124. | ||
Su N, Wang Y, Qian M, Deng M. Combinatorial regulation of transcription factors and microRNAs. BMC Syst Biol. 2010;4:150. | ||
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. | ||
Schmitt MJ, Margue C, Behrmann I, Kreis S. MiRNA-29: a microRNA family with tumor-suppressing and immune-modulating properties. Curr Mol Med. 2013;13(4):572–585. | ||
Liu NK, Xu XM, Xm X. MicroRNA in central nervous system trauma and degenerative disorders. Physiol Genomics. 2011;43(10):571–580. | ||
Nelson PT, Wang WX, Rajeev BW, Micrornas RBW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18(1):130–138. | ||
Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39(3):959–966. | ||
Ji Q, Ji Y, Peng J, et al. Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One. 2016;11(9):e0163645. | ||
Mahlamäki EH, Bärlund M, Tanner M, et al. Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer. 2002;35(4):353–358. | ||
Varis A, Wolf M, Monni O, et al. Targets of gene amplification and overexpression at 17q in gastric cancer. Cancer Res. 2002;62(9):2625–2629. | ||
Li Y, Wang J, Dai X, et al. miR-451 regulates FoxO3 nuclear accumulation through Ywhaz in human colorectal cancer. Am J Transl Res. 2015;7(12):2775–2785. | ||
Gu X, Li JY, Guo J, Li PS, Zhang WH. Influence of MiR-451 on drug resistances of paclitaxel-resistant breast cancer cell line. Med Sci Monit. 2015;21:3291–3297. | ||
Liu K, Tian H, Zhang Y, Zhao H, Ma K. miR-451 selectively increases sensitivity to cisplatin in ERCC1-high non-small cell lung cancer cells. J Cell Biochem. 2018;06:29315773. | ||
Blech-Hermoni Y, Stillwagon SJ, Ladd AN. Diversity and conservation of CELF1 and CELF2 RNA and protein expression patterns during embryonic development. Dev Dyn. 2013;242(6):767–777. | ||
Ajith S, Gazzara MR, Cole BS, et al. Position-dependent activity of CELF2 in the regulation of splicing and implications for signal-responsive regulation in T cells. RNA Biol. 2016;13(6):569–581. | ||
Fan B, Jiao BH, Fan FS, et al. Downregulation of miR-95-3p inhibits proliferation, and invasion promoting apoptosis of glioma cells by targeting CELF2. Int J Oncol. 2015;47(3):1025–1033. | ||
Miyazaki Y, Adachi H, Katsuno M, et al. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18(7):1136–1141. | ||
Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila). 2009;2(9):807–813. | ||
Godlewski J, Bronisz A, Nowicki MO, Chiocca EA, Lawler S. microRNA-451: a conditional switch controlling glioma cell proliferation and migration. Cell Cycle. 2010;9(14):2814–2820. | ||
Zhang X, Wang X, Zhu H, et al. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49(5):841–850. | ||
Xie J, Hu X, Yi C, Hu G, Zhou X, Jiang H. MicroRNA-451 protects against cardiomyocyte anoxia/reoxygenation injury by inhibiting high mobility groupbox1 expression. Mol Med Rep. 2016;13(6):5335–5341. | ||
Minta M, Radko L, Stypuła-Trębas S, Żmudzki J. Cytotoxic effects of the synthetic oestrogens and androgens on Balb/c 3T3 and HepG2 cells. Bull Vet Inst Pulawy. 2014;58(4):613–620. | ||
Mirzaee M, Moieni A, Ghanati F. Effects of drought stress on the lipid peroxidation and antioxidant enzyme activities in two canola (Brassica napus L. cultivars). J Agri Sci Tec. 2013;15(3):593–602. | ||
Chen H, Chow PH, Cheng SK, Cheung AL, Cheng LY, O WS. Male genital tract antioxidant enzymes: their source, function in the female, and ability to preserve sperm DNA integrity in the golden hamster. J Androl. 2003;24(5):704–711. | ||
Ramalingam S, Ramamoorthy P, Subramaniam D, Anant S. Reduced expression of RNA binding protein CELF2, a putative tumor suppressor gene in colon cancer. Immunogastroenterology. 2012;1(1):27–33. | ||
Office U. Department of the Treasury, Internal Revenue Service; Department of Labor, Employee Benefits Security Administration; Department of Health and Human Services: Summary of Benefits and Coverage and Uniform Glossary. P Natl Acad Sci. 2015;112(17):2139–2148. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.