Back to Journals » Cancer Management and Research » Volume 12
MicroRNA-431-5p Inhibits the Tumorigenesis of Osteosarcoma Through Targeting PANX3
Authors Sun S, Fu L, Wang G, Wang J, Xu L
Received 27 April 2020
Accepted for publication 13 August 2020
Published 8 September 2020 Volume 2020:12 Pages 8159—8169
DOI https://doi.org/10.2147/CMAR.S260149
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Ahmet Emre Eşkazan
Shengliang Sun,1,* Lei Fu,1,* Gen Wang,1 Jianli Wang,1 Liping Xu2
1Department of Orthopedic Trauma, The 89th Army Hospital of the Chinese People’s Liberation Army, Weifang, Shandong, People’s Republic of China; 2Department of Oncology, The 89th Army Hospital of the Chinese People’s Liberation Army, Weifang, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liping Xu Tel +86 536-5019053
Email [email protected]
Purpose: This study aimed to evaluate the regulatory role of miR-431-5p on the tumorigenesis of osteosarcoma (OS) and the underlying mechanism involving pannexin 3 (PANX3).
Methods: qRT-PCR was applied to measure the expression of miR-431-5p in OS tissues and cells. PANX3 and miR-431-5p were overexpressed in U2OS and HOS cells. The cell viability and apoptosis were determined by MTT and FITC/PI double staining assay, respectively. Transwell assay was performed to detect cell migration and invasion. The protein expression of cleave-caspase-3 and MMP-2/-9 was detected by Western blot. The target relationship between miR-431-5p and PANX3 was predicated by ENCORI and identified by DLR assay. The anti-tumor effect of miR-431-5p was further analyzed in a xenograft tumor model in mice.
Results: MiR-431-5p expression was down-regulated in OS tissues and negatively correlated with lymph node metastasis and TNM stage. Over-expression of miR-431-5p induced cell apoptosis, inhibited cell proliferation, migration and invasion, up-regulated cleave-caspase-3, and down-regulated MMP-2 and -9 in OS cells. Over-expression of miR-431-5p also inhibited the growth of tumor xenografts in mice. In addition, PANX3 was a target of miR-431-5p. Over-expression of PANX3 reversed the anti-tumor effect of miR-431-5p mimics on U2OS and HOS cells.
Conclusion: Up-regulation of miR-431-5p suppressed the tumorigenesis of OS via targeting PANX3.
Keywords: osteosarcoma, microRNA-431-5p, pannexin 3, proliferation, migration
Introduction
Osteosarcoma (OS) is a severe tumor in bone that commonly occurs in adolescents with a high incidence at the age of 15–19 years.1,2 Surgical resection, radiotherapy, and chemotherapy are common therapeutic strategies for OS.3 Although substantial improvements have been made on the treatment of OS, the outcomes of OS cases, especially those with metastasis and recurrence remain poor.4 Therefore, it is urgently needed to search for novel therapeutic targets for OS.
The emerging role of microRNAs (miRNAs) in controlling key pathways implicated in tumorigenesis makes their use as a powerful novel tool for the early detection, risk assessment and prognosis, as well as for the development of innovative anticancer therapies.5 Massive miRNAs are involved in the tumorigenesis of OS.6 A previous study based on gene expression profiling has shown that seven miRNAs are associated with OS, including miR-422a, -145, -194, -93, -125b, -193a-3p, and -140-5p.7 Some miRNAs have been proved to be down-regulated in OS, such as miR-30a,8 -33b,9 -101,10 -379,11 -493-5p,12 -567,13 and -638.14 These miRNAs exert valuable anti-tumor effect on OS through regulating diverse cellular processes.11–13 For example, miR-379 and miR-567 can both suppress the proliferation, invasion, and migration of U2OS and MG-63 cells.11,13 Over-expression of miR-493-5p induces apoptosis and suppresses the viability, migration, and invasion of U2OS cells.12 MiR-431 is a specific miRNA that is involved in cancer development. MiR-431 is down-regulated in colorectal cancer tissues, and the up-regulation of miR-431 suppresses the migration of HCT116 and SW480 cells.15 Sun et al have shown that miR-431 is down-regulated in HCC tissues, and miR-431 over-expression suppresses the invasion and migration of HCCLM3 cells.16 Notably, a miRNA profile of OS patients has found that miR-431-5p is down-regulated in OS.17 However, the regulatory role of miR-431-5p on OS remains unknown.
The regulatory effects of miRNAs on tumor cells are mainly achieved by modulating specific target genes.18,19 Many regulatory interactions between miRNAs and target genes in OS have been found by previous studies, such as miR-101 and ROCK1,10 miR-379 and EIF4G2,11 miR-493-5p and KLF5,12 and miR-567 and FGF5.13 Pannexins (PANX1, 2 and 3) are a family of single membrane channel-forming glycoproteins that mainly function in cellular communication and autocrine/paracrine signaling.20 PANX3 plays a key role in osteoblast differentiation and osteogenesis.21 Romano et al have shown that PANX3 is up-regulated in an axillary sweat gland carcinoma with osteosarcomatous transformation, and high expression of PANX3 strongly predicts OS.22 A whole transcriptome analysis has revealed that PANX3 is up-regulated in OS tissues.23 However, the regulatory role of PANX3 and its regulatory relationship with miR-431-5p on OS remain unclear.
Here, the expression of miR-431-5p in OS tissues and cells was determined. The regulatory role of miR-431-5p was evaluated on the proliferation, apoptosis, migration, and invasion of OS cells, and on the growth of tumor xenografts in mice. The regulatory relationship between PANX3 and miR-431-5p was further investigated. Our study may reveal a promising target for the treatment of OS.
Materials and Methods
Tumor Samples
A total of 62 OS patients (33 male and 29 female; 21–78 years old) were screened from our hospital between January 2016 and January 2019. OS tissues and adjacent normal tissues were collected by surgical resection and were confirmed histopathologically. Patients had not received chemotherapy and/or radiotherapy before resection. This study was approved by the Ethics Committee of The 89th Army Hospital of the Chinese People’s Liberation Army in accordance with the Declaration of Helsinki, and informed consents were obtained from all subjects.
Cell Culture
Four OS cell lines (U2OS, SaOS-2, SJSA-1 and HOS) and one normal human osteoblast cell line (hFOB 1.19) were purchased from BeiNa Chuanglian Biotechnology Research Institute (Beijing, China). Cells were cultured in DMEM containing 10% FBS at 37°C with 5% CO2.
Cell Transfection
The miR-431-5p mimics, mimics negative control (mimics-NC), pcDNA3.1-PANX3 (pc-PANX3), and empty pcDNA3.1 (pcDNA3.1) were obtained from GenePharma (Shanghai, China). Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA, USA) was used for cell transfection. Cells were randomly divided into Blank (cells without transfection), miR-431-5p mimics, mimics-NC, mimics-NC + pcDNA3.1, mimics-NC + pc-PANX3, miR-431-5p mimics + pcDNA3.1, and miR-431-5p mimics + pc-PANX3 groups. After 48 h of transfection, the transfected cells were used for the subsequent assays.
MTT Assay
Cells (1 × 104 cells/well) were seeded into 96-well plates and cultured for 0, 24, 48 and 72 h, respectively. After 4 h of incubation with MTT, DMSO was added to dissolve the formazan crystals. The optical density (OD) at 490 nm was detected by a Microplate Reader.
FITC/PI Double Staining Assay
Cell apoptosis was detected using an Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) kit (Thermo Fisher Scientific). Briefly, cells (1 × 105 cells/well) were seeded into 6-well plates and incubated with Annexin V-FITC and PI in the dark for 15 min. The cell apoptosis was monitored by a flow cytometry (BD Biosciences, San Jose, CA, USA) using cytexpert software (V2.0). The apoptotic cells (%) were calculated as cells in the lower right quadrant (early apoptotic cells) + upper right quadrant (late apoptotic cells).
Transwell Assay
Cells (1 × 105 cells) in FBS-free DMEM were seeded into the upper chamber and DMEM containing 10% FBS was added into the lower chamber. After 24 h of culturing, cells on the lower chamber were stained with 0.1% crystal violet for 20 min. The stained cells (migration cells) were counted under a microscope (Olympus, Tokyo, Japan) at five randomly selected fields. The upper chamber was pre-coated with Matrigel for detecting cell invasion.
qRT-PCR
Total RNAs were extracted from tissues and cells using TRIZOL and were then transcribed into cDNA. qRT-PCR was performed on a real-time PCR system (ABI 7500) using the following program: 94°C for 5 min, 35 cycles of 94°C for 10 s, 56°C for 30 s and 72°C for 30 s. The relative expression level of specific genes was quantified using the 2−ΔΔCt method. The internal controls were GAPDH and U6. The primers were synthesized by Sangon Biotech Co, Ltd (Shanghai, China) and the sequences are shown in Table 1.
 |
Table 1 The Primers Used in qRT-PCR |
Western Blot
The proteins were isolated by lysing cells in RIPA lysis buffer. The proteins were then separated by 10% SDS-PAGE and transferred onto PVDF membrane. After 2 h of blocking with 5% skim milk, the membrane was incubated with primary antibody (anti-PANX3, -cleave-caspase-3, -MMP-2, -MMP-9, -GAPDH; 1:1000, Thermo Fisher Scientific) at 4°C for 12 h. The membrane was then incubated with HRP-conjugated secondary antibody (1:1000, Thermo Fisher Scientific) for 2 h at 25°C. The bands were visualized using ECL kit and quantified by a Gel Imaging System. GAPDH was used as the internal control.
Dual-Luciferase Reporter (DLR) Assay
A binding site of miR-431-5p at 3ʹ-UTR of PANX3 was predicted by ENCORI. The fragments of PANX3 containing the predicted binding site (Wt) and mutant site (Mut) were synthesized and inserted into pGL3-promoter vector (PANX3-Wt/PANX3-Mut). U2OS and HOS cells were co-transfected with miR-431-5p mimics/mimics-NC and PANX3-Wt/PANX3-Mut using Lipofectamine 3000 for 48 h. After transfection, cells were lysed in Lysis Buffer. Luciferase Assay Reagent was added into the lysate and the intensity of Fireny luciferase was detected by a Microplate Reader. Subsequently, Stop & Glo Reagent was added and the intensity of Renilla luciferase was detected. The relative luciferase activity was calculated as the ratio of Fireny luciferase/Renilla luciferase.
Subcutaneous Tumor Xenografts in Mice
Nude mice (BALB/c, male, four-week-old, N = 18) were obtained from SLAC (Shanghai, China) and maintained in a controlled environment at 23–25°C, 50–60% humidity, and 12-h light/dark cycle with free access to food and water. Mice were randomly divided into Blank, mimics-NC, and miR-431-5p mimics groups (N = 6 each group). U2OS cells (1 × 107 cells) transfected with mimics-NC or miR-431-5p mimics were subcutaneously injected into the right flank of mice. Mice injected with non-transfected U2OS cells were considered as the Blank group. The tumor volume was measured every 5 days after the injection using a caliper and calculated using the following formula: (A×B2)/2 (A, the longest diameter; B, the shortest diameter). At the 30th day post-injection, mice were anesthetized with pentobarbital sodium (60 mg/kg) and killed by cervical dislocation. The tumor xenografts were removed and weighed. All animal experiments were approved by the Ethics Committee of The 89th Army Hospital of the Chinese people’s Liberation Army in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA).
Statistical Analysis
The data were expressed as the mean ± standard deviation and analyzed using SPSS 22.0 software. The differences among multi-groups were determined by one-way ANOVA followed by Tukey’s post hoc test. The differences between two groups were determined by Student’s t test. The correlation between miR-431-5p and PANX3 expression was analyzed by Pearson correlation test. A p < 0.05 was considered to be significantly different.
Results
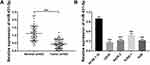
The miR-431-5p Was Down-Regulated in OS
The expression of miR-431-5p was determined in OS tissues and cells. MiR-431-5p expression in OS tissues was significantly lower than that in adjacent normal tissues (P < 0.001, Figure 1A). MiR-431-5p expression in OS cell lines (U2OS, SaOS-2, SJSA-1 and HOS) was also significantly lower than that in hFOB 1.19 cells (P < 0.001, Figure 1B). We then analyzed the correlation between miR-431-5p expression and clinicopathological parameters of OS patients. The results showed that miR-431-5p expression was negatively correlated with the TNM stage and lymph node metastasis (P < 0.05) (Table 2).
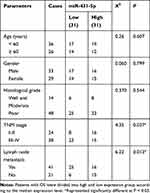 |
Table 2 The Correlation Between the Expression of miR-431-5p and the Clinicopathological Parameters of Patients with OS |
Over-Expression of miR-431-5p Inhibited the Malignant Behavior of OS Cells
MiR-431-5p was over-expressed in U2OS and HOS cells by the transfection of miR-431-5p mimics (P < 0.001, Figure 2A). The cell viability in the miR-431-5p mimics group was significantly decreased compared with that in the mimics-NC group (P < 0.05, Figure 2B). On the contrary, the apoptosis rate in the miR-431-5p mimics group was significantly increased compared with that in the mimics-NC group (P < 0.001, Figure 2C). Transwell assay showed that the transfection of miR-431-5p mimics promoted the migration and invasion of U2OS and HOS cells (P < 0.001, Figure 2D and E).
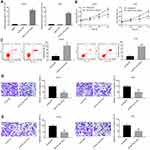
PANX3 Was a Target of miR-431-5p
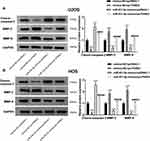
A binding site of miR-431-5p was predicted at 3ʹ-UTR of PANX3 by ENCORI (Figure 3A). The target relationship between miR-431-5p and PANX3 was further confirmed by DLR assay (P < 0.01, Figure 3B). The mRNA and protein expression of PANX3 in OS cell lines (U2OS, SaOS-2, SJSA-1 and HOS) were higher than those in normal human osteoblast cells (hFOB 1.19) (P < 0.001, Figure 3C). MiR-431-5p mimics significantly decreased the mRNA and protein expression of PANX3 in U2OS and HOS cells (P < 0.001, Figure 3D and E). In addition, the mRNA expression of PANX3 in OS tissues was higher than that in adjacent normal tissues (P < 0.001, Figure 3F). Pearson correlation test showed that PANX3 expression was negatively correlated with miR-431-5p expression in OS tissues (P = 0.0085, Figure 3G).
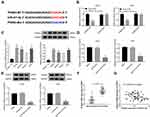
Over-Expression of PANX3 Reversed the Anti-Tumor Effect of miR-431-5p Mimics on OS Cells
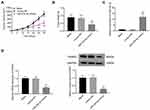
PANX3 was over-expressed in U2OS and HOS cells. The protein expression of PANX3 in the mimics-NC + pc-PANX3 group was increased compared with that in the mimics-NC + pcDNA3.1 group (P < 0.01). pc-PANX3 reversed the inhibiting effect of miR-431-5p mimics on the protein expression of PANX3 in U2OS and HOS cells (P < 0.01, Figure 4A). In addition, the cell viability, migration, and invasion were enhanced in the mimics-NC + pc-PANX3 group (P < 0.05). pc-PANX3 reversed the inhibiting effects of miR-431-5p mimics on the viability, migration, and invasion of U2OS and HOS cells (P < 0.01, Figure 4B, D and E). On the contrary, pc-PANX3 reduced the apoptosis rate of U2OS and HOS cells, and reversed the promoting effect of miR-431-5p mimics on cell apoptosis (P < 0.01, Figure 4C).
Over-Expression of miR-431-5p Up-Regulated Cleave-Caspase-3 and Down-Regulated MMP-2 and -9 in OS Cells via Targeting PANX3
In order to further identify the regulatory role of miR-431-5p on the apoptosis and metastasis of OS cells, the expression of apoptotic marker cleave-caspase-3 and metastasis markers MMP-2 and -9 were detected. As shown in Figure 5A and B, the protein expression of cleave-caspase-3 was significantly increased by the transfection of miR-431-5p mimics in U2OS and HOS cells (P < 0.001). Over-expression of PANX3 significantly reversed the promoting effect of miR-431-5p mimics on the protein expression of cleave-caspase-3 (P < 0.01). In addition, the protein expression of MMP-2 and -9 was significantly decreased by the transfection of miR-431-5p mimics in U2OS and HOS cells (P < 0.001). Over-expression of PANX3 significantly reversed the inhibiting effect of miR-431-5p mimics on the protein expression of MMP-2 and -9 (P < 0.001) (Figure 5A and B).
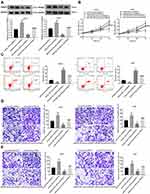
Over-Expression of miR-431-5p Inhibited the Growth of Tumor Xenografts in Mice
Subcutaneous tumor xenografts in mice were used to identify the anti-tumor role of miR-431-5p in vivo. The tumor volumes in the miR-431-5p mimics group were decreased compared with that in the Blank group beginning from the 15th day (P < 0.05, Figure 6A). At the 30th day post-injection, the tumor weight in the miR-431-5p mimics group was lower than that in the Blank group (P < 0.01, Figure 6B). Mimics-NC did not significantly influence the growth of tumor xenografts in mice. In addition, miR-431-5p mimics significantly increased the expression of miR-431-5p and decreased the expression of PANX3 in tumor xenografts (P < 0.01, Figure 6C and D).
Discussion
MiRNAs are key regulators in the tumorigenesis of OS. Some miRNAs are down-regulated in OS, such as miR-133a,24 -137,25 -150,26 -223,27 -505,28 and -564.29 In consistent with these miRNAs, miR-431-5p was also down-regulated in OS tissues and cells in this study. Our findings indicate that miR-431-5p may be a tumor suppressor in OS. The down-regulation of miRNAs is usually associated with the clinical characteristics of OS patients. Li et al and Qu et al have shown that miR-505 and miR-150 are both negatively associated with TNM stage and metastasis status in patients with OS.26,28 Dong et al have shown that miR-223 is negatively related to distant metastasis, advanced stage, and poor survival time in patients with OS.27 Here, TNM stage and lymph node metastasis were negatively correlated with miR-431-5p expression in OS tumors. The clinical predictive value of miR-223 on OS is consistent with the above miRNAs. MiR-223 may be a potential diagnostic and prognostic marker for OS.
MiRNAs play key roles in the occurrence and progression of OS by regulating cellular processes. For example, miR-567, -192, and -152 can inhibit the proliferation, migration, and invasion of MG-63 and/or U2OS cells.13,30,31 Here, miR-431-5p was over-expressed in OS cells to identify the regulatory role of miR-431-5p on OS cells. The up-regulation of miR-431-5p promoted the apoptosis and inhibited the proliferation, migration, and invasion of U2OS and HOS cells. Our findings are similar to the above previous researches, and illustrate that miR-431-5p acts as a tumor suppressor in OS cells. In addition, we also found that the up-regulation of miR-431-5p up-regulated apoptotic marker cleave-caspase-3 and down-regulated metastasis markers MMP-2 and -9 in U2OS and HOS cells. These results further illustrate that miR-431-5p can promote the apoptosis and inhibit the metastasis of OS cells. The anti-tumor effect of miR-431-5p on OS is not only limited at the cellular level but also reflected in animal models. In this study, subcutaneous tumor xenografts were established in mice to identify the anti-tumor role of miR-431-5p in vivo. The results showed that the tumor volume and weight were decreased by the miR-431-5p mimics in mice. These findings indicate that the over-expression of miR-431-5p can inhibit the tumor growth in vivo.
The anti-tumor effects of miRNAs on OS are always achieved by modulating specific target genes. In this study, PANX3 was identified as a target of miR-431-5p, and miR-431-5p mimics down-regulated PANX3 in U2OS and HOS cells. These findings indicate that miR-431-5p negatively regulated its target gene PANX3 in OS cells. PANX3 is a member of the gap junction PANX family that plays an important role in coordinating tissue homeostasis.32 Previous studies have revealed that some PANX members are involved in the processes of multi-types of cancers. For example, silencing of PANX1 suppresses the proliferation of malignant glioma cells.33 Over-expression of PANX1 inhibits the spheroid formation and growth of rhabdomyosarcoma cells.34 Loss of PANX1 attenuates the melanoma progression by reversion to a melanocytic phenotype.35 In this study, over-expression of PANX3 inhibited apoptosis and promoted the proliferation, migration, and invasion of U2OS and HOS cells. The function of PANX3 in OS is similar to that of PANX1 in glioma and melanoma. We speculate that miR-431-5p may inhibit the tumorigenesis of OS through down-regulating PANX3. This speculation was subsequently confirmed by the fact that over-expression of PANX3 significantly reversed the anti-tumor effect of miR-431-5p mimics on OS cells.
Conclusions
In conclusion, miR-431-5p was down-regulated in OS tissues and negatively correlated with lymph node metastasis and TNM stage in OS patients. Over-expression of miR-431-5p induced the apoptosis and inhibited the proliferation, migration, and invasion of OS cells through targeting PANX3. Over-expression of miR-431-5p also inhibited the growth of tumor xenografts in mice. MiR-431-5p and its target PANX3 may be therapeutic targets for OS. However, further studies on the upstream regulatory factors and other target genes of miR-431-5p are still needed.
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of The 89th Army Hospital of the Chinese People’s Liberation Army in accordance with the Declaration of Helsinki, and informed consents were obtained from all subjects. All animal experiments were approved by the Ethics Committee of The 89th Army Hospital of the Chinese people’s Liberation Army in accordance with the Guide for the Care and Use of Laboratory Animals (eighth edition, 2011, National Institutes of Health, USA).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest to disclose for this work.
References
1. Oryan A, Alidadi S, Moshiri A. Osteosarcoma: current concepts, challenges and future directions. Curr Orthop Pract. 2015;26(2):181–198. doi:10.1097/BCO.0000000000000199
2. Mirabello L, Troisi R, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the surveillance, epidemiology, and end results program. Cancer. 2009;115(7):1531–1543. doi:10.1002/cncr.24121
3. Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2017;18(1):39. doi:10.1080/14737140.2018.1413939
4. Sayles LC, Breese MR, Koehne AL, et al. Genome-informed targeted therapy for osteosarcoma. Cancer Discov. 2019;9(1):46–63. doi:10.1158/2159-8290.CD-17-1152
5. Gulino R, Forte S, Parenti R, Memeo L, Gulisano M. MicroRNA and pediatric tumors: future perspectives. Acta Histochem. 2015;117(4–5):339–354. doi:10.1016/j.acthis.2015.02.007
6. Calderón SAL, Garbutt C, Kim J, et al. Clinical and molecular analysis of pathologic fracture-associated osteosarcoma: microRNA profile is different and correlates with prognosis. Clin Orthop Relat Res. 2019;477(9):2114–2126. doi:10.1097/CORR.0000000000000867
7. Fan H, Lu S, Wang S, Zhang S. Identification of critical genes associated with human osteosarcoma metastasis based on integrated gene expression profiling. Mol Med Rep. 2019;20(2):915–930. doi:10.3892/mmr.2019.10323
8. Yuan T, Wang J, Wei X, Wang G, Xu N, Fan L. MicroRNA-30a inhibits proliferation and metastasis of osteosarcoma cells by modulating autophagy. J Biobased Mater Bioenergy. 2016;10(4):265–271. doi:10.1166/jbmb.2016.1588
9. Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol Res. 2017;25(3):397–405. doi:10.3727/096504016X14743337535446
10. Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 2017;7(1):88–97.
11. Xie X, Li YS, Xiao WF, et al. MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targetting EIF4G2. Biosci Rep. 2017;37(3). doi:10.1042/BSR20160542.
12. Zhang Z, Luo G, Yu C, Yu G, Jiang R, Shi X. MicroRNA-493-5p inhibits proliferation and metastasis of osteosarcoma cells by targeting Kruppel-like factor 5. J Cell Physiol. 2019;234(8):13525–13533. doi:10.1002/jcp.28030
13. Liu D, Zhang C, Li X, Zhang H, Wan A. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI J. 2018;17:102–112. doi:10.17179/excli2017-932
14. Wang XX, Liu J, Tang YM, Hong L, Tan GH. MicroRNA-638 inhibits cell proliferation by targeting suppress PIM1 expression in human osteosarcoma. Tumour Biol. 2017;37(12):1–9. doi:10.1007/s13277-015-4142-3
15. Su WB, Liu ZY. MiR-431 inhibits colorectal cancer cell invasion via repressing CUL4B. Eur Rev Med Pharmacol Sci. 2018;22(10):3047–3052. doi:10.26355/eurrev_201805_15062
16. Sun K, Zeng T, Huang D, et al. MicroRNA-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the ZEB1-mediated epithelial–mesenchymal transition. FEBS Open Bio. 2015;5(1):900–907. doi:10.1016/j.fob.2015.11.001
17. Andersen GB, Knudsen A, Hager H, Hansen LL, Tost J. miRNA profiling identifies deregulated miRNAs associated with osteosarcoma development and time to metastasis in two large cohorts. Mol Oncol. 2018;12(1):114–131. doi:10.1002/1878-0261.12154
18. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
19. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
20. Jiang JX, Penuela S. Connexin and pannexin channels in cancer. BMC Cell Biol. 2016;17(S1):S12. doi:10.1186/s12860-016-0094-8
21. Ishikawa M, Yamada Y. The role of pannexin 3 in bone biology. J Dent Res. 2017;96(4):372–379. doi:10.1177/0022034516678203
22. Romano RC, Gardner JM, Shalin SC, et al. High relative expression of Pannexin 3 (PANX3) in an axillary sweat gland carcinoma with osteosarcomatous transformation. Am J Dermatopathol. 2016;38(11):846–851. doi:10.1097/DAD.0000000000000583
23. Ho XD, Phung P, LeV Q, et al. Whole transcriptome analysis identifies differentially regulated networks between osteosarcoma and normal bone samples. Exp Biol Med. 2017;242(18):1802–1811. doi:10.1177/1535370217736512
24. Ji F, Zhang H, Wang Y, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56(1):220–226. doi:10.1016/j.bone.2013.05.020
25. Li ZM, Zhang HY, Wang YX, Wang WB. MicroRNA-137 is downregulated in human osteosarcoma and regulates cell proliferation and migration through targeting FXYD6. J Drug Target. 2016;24(2):102–110. doi:10.3109/1061186X.2015.1057149
26. Li CH, Yu TB, Qiu HW, Zhao X, Zhou CL, Qi C. miR-150 is downregulated in osteosarcoma and suppresses cell proliferation, migration and invasion by targeting ROCK1. Oncol Lett. 2017;13(4):2191–2197. doi:10.3892/ol.2017.5709
27. Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5(2):74–79. doi:10.1016/j.jbo.2016.05.001
28. Liu YJ, Li W, Chang F, Liu JN, Lin JX, Chen DX. MicroRNA-505 is downregulated in human osteosarcoma and regulates cell proliferation, migration and invasion. Oncol Rep. 2018;39(2):491–500. doi:10.3892/or.2017.6142
29. Ru N, Zhang F, Liang J, et al. MiR-564 is down-regulated in osteosarcoma and inhibits the proliferation of osteosarcoma cells via targeting Akt. Gene. 2018;645:163–169. doi:10.1016/j.gene.2017.12.028
30. Shang G, Mi Y, Mei Y, Wang G, Wang Y. MicroRNA-192 inhibits the proliferation, migration and invasion of osteosarcoma cells and promotes apoptosis by targeting matrix metalloproteinase-11. Oncol Res. 2018;15(5):7265–7272.
31. Ma C, Han J, Dong D, Wang N. MicroRNA-152 suppresses human osteosarcoma cell proliferation and invasion by targeting E2F transcription factor 3. Oncol Res. 2018. doi:10.3727/096504017X15021536183535
32. Makarenkova HP, Shestopalov VI. The role of pannexin hemichannels in inflammation and regeneration. Front Physiol. 2014;5:63. doi:10.3389/fphys.2014.00063
33. Wei L, Yang X, Shi X, Chen Y. Pannexin-1 silencing inhibits the proliferation of U87-MG cells. Mol Med Rep. 2015;11(5):3487–3492. doi:10.3892/mmr.2015.3169
34. Xiang X, Langlois S, St-Pierre ME, Barre J, Cowan KN. Abstract 1276: pannexin 1 regulates rhabdomyosarcoma tumor growth: a potential novel therapeutic target. Cancer Res. 2016;76(14 Supplement):1276.
35. Penuela S, Gyenis L, Ablack A, et al. Loss of pannexin 1 attenuates melanoma progression by reversion to a melanocytic phenotype. J Biol Chem. 2012;287(34):29184–29193. doi:10.1074/jbc.M112.377176
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.